Amplite® Fluorimetric Sphingomyelin Assay Kit *Red Fluorescence*
| Price | |
| Catalog Number | |
| Unit Size | |
| Quantity |
| Telephone | 1-800-990-8053 |
| Fax | 1-800-609-2943 |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Shipping | Standard overnight for United States, inquire for international |
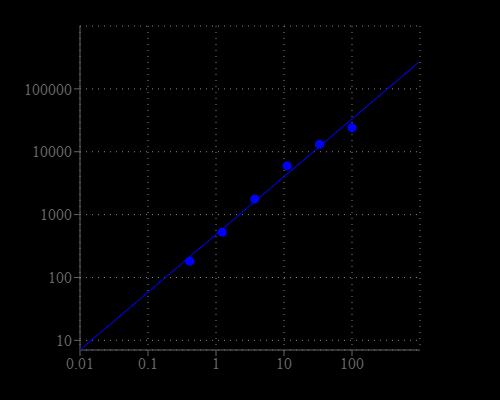
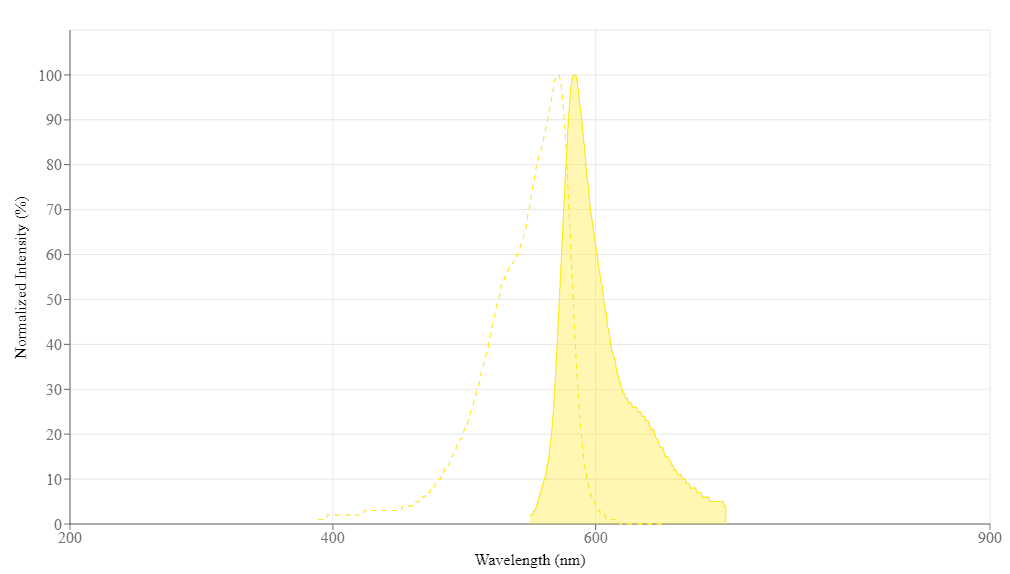
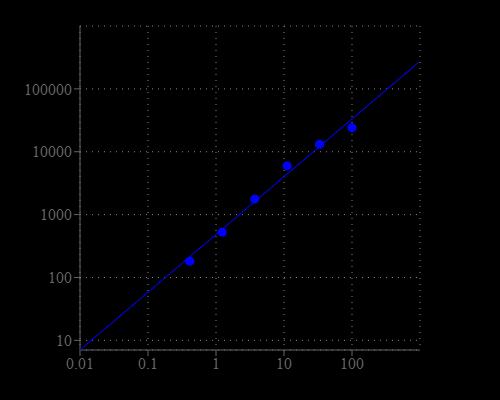
| Excitation (nm) | 571 |
| Emission (nm) | 584 |
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
| Overview |
Excitation (nm) 571 | Emission (nm) 584 |
Platform
Fluorescence microplate reader
| Excitation | 540 nm |
| Emission | 590 nm |
| Cutoff | 570 nm |
| Recommended plate | Solid black |
Components
Example protocol
AT A GLANCE
Protocol summary
- Prepare SMase working solution (50 µL)
- Add sphingomyelin standards or test samples (50 µL)
- Incubate at 37 °C for 1 - 2 hours
- Add sphingomyelin assay mixture (50 µL)
- Incubate at RT for 0.5 - 2 hours
- Monitor fluorescence intensity at Ex/Em = 540/590 nm (cut off at 570 nm)
Important notes
Thaw kit components at room temperature before starting your experiment.
PREPARATION OF STOCK SOLUTION
1. SMase stock solution (100X):
Add 50 µL of PBS with 0.1% BSA into the vial of Sphingomyelinase (Component B) to make SMase stock solution (100X).
2. Amplite™ Red stock solution (200X):
Add 80 µL of DMSO (Component G) into the vial of Amplite™ Red (Component C) to make 200X Amplite™ Red stock solution. Keep from light. Note: The Amplite™ Red is unstable in the presence of thiols (such as DTT and 2-mercaptoethanol). The final concentration of DTT or 2-mercaptoethanol in the reaction should be lower than 10 µM. Amplite™ Red is also unstable at high pH (>8.5). The reactions should be performed at pH 7 - 8. pH 7.4 is recommended for the assay buffer.
PREPARATION OF STANDARD SOLUTION
For convenience, use the Serial Dilution Planner: https://www.aatbio.com/tools/serial-dilution/13625
Add 2 µL of 50 mM Sphingomyelin (Component F) into 1000 µL of SMase Reaction Buffer (Component D) to get a 100 µM Sphingomyelin standard solution (SM7). Take 100 µM Sphingomyelin standard solution to perform 1:3 serial dilutions to get serially diluted sphingomyelin standards (SM6 - SM1).
PREPARATION OF WORKING SOLUTION
1. Sphingomyelinase (SMase) working solution:
Prepare SMase working solution by adding the whole content (50 µL) of 100X SMase stock solution into 5 mL of SMase Reaction Buffer (Component D) and mix well. Note: The SMase working solution should be used promptly.
2. Sphingomyelin working solution:
Add the whole content (5 mL) of Assay Buffer (Component E) into the bottle of Enzyme Mix (Component A) and mix well. Add 25 µL 200X Amplite™ Red stock solution into the bottle of Enzyme Mix solution to make the sphingomyelin assay mixture before starting the assay. Note: The sphingomyelin assay mixture should be used promptly and kept from light; longer storage is likely to cause high assay background.
For guidelines on cell sample preparation, please visit
https://www.aatbio.com/resources/guides/cell-sample-preparation.html
SAMPLE EXPERIMENTAL PROTOCOL
Table 1. Layout of sphingomyelin standards and test samples in a solid black 96-well microplate. SM = Sphingomyelin Standards (SM1 - SM7, 0.1 to 100 µM), BL = Blank Control, TS = Test Samples.
| BL | BL | TS | TS |
| SM1 | SM1 | ... | ... |
| SM2 | SM2 | ... | ... |
| SM3 | SM3 | ||
| SM4 | SM4 | ||
| SM5 | SM5 | ||
| SM6 | SM6 | ||
| SM7 | SM7 |
Table 2. Reagent composition for each well.
| Well | Volume | Reagent |
| SM1 - SM7 | 50 µL | Serial Dilutions (0.1 to 100 µM) |
| BL | 50 µL | SMase Reaction Buffer |
| TS | 50 µL | test sample |
- Prepare sphingomyelin standards (SM), blank controls (BL), and test samples (TS) according to the layout provided in Tables 1 and 2. For a 384-well plate, use 25 µL of reagent per well instead of 50 µL. Note: Treat your cells or tissue samples as desired.
- Add 50 µL of SMase working solution to each well of sphingomyelin standard, blank control, and test samples to make the total sphingomyelin assay volume of 100 µL/well. For a 384-well plate, add 25 µL of SMase working solution into each well instead, for a total volume of 50 µL/well.
- Incubate the reaction mixture at 37°C for 1 - 2 hours.
- Add 50 µL of sphingomyelin assay mixture to each well of sphingomyelin standard, blank control, and test samples to make the total sphingomyelin assay volume of 150 µL/well. For a 384-well plate, add 25 µL of sphingomyelin assay mixture into each well instead, for a total volume of 75 µL/well.
- Incubate the reaction mixture for 1 - 2 hours at room temperature (protected from light).
- Monitor the fluorescence increase with a fluorescence microplate reader at Ex/Em = 540/590 nm (cut off at 570 nm).
Images
Citations
Authors: Yagi, Mayuko and Hama, Minami and Ichii, Sayaka and Nakashima, Yurie and Kanbayashi, Daiki and Kurata, Takako and Yusa, Kosuke and Komano, Jun
Journal: iScience (2023)
Authors: Wang, Yang and Chen, Yan-Yan and Gao, Gui-Bin and Zheng, Yang-Han and Yu, Nan-Nan and Ouyang, Lan and Gao, Xuejuan and Li, Nan and Wen, Shi-Yuan and Huang, Shangjia and others,
Journal: Molecular Therapy (2023)
Authors: Prymas, Kamila and Swiatkowska, Anna and Traczyk, Gabriela and Ziemlińska, Ewelina and Dziewulska, Anna and Ciesielska, Anna and Kwiatkowska, Katarzyna
Journal: Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids (2019): 158549
Authors: Su, Yu-Ting and Meng, Xing-Xing and Zhang, Xi and Guo, Yi-Bin and Zhang, Hai-Jun and Cheng, Yao-Ping and Xie, Xiao-Ping and Chang, Yao-Ming and Bao, Jun-Xiang
Journal: The Anatomical Record (2017)
Authors: Li, Lin and Niu, Huanmin and Sun, Bin and Xiao, Yanan and Li, Wei and Yuan, Huiqing and Lou, Hongxiang
Journal: Toxicology and Applied Pharmacology (2016): 175--184
Authors: Mashhadi Akbar Boojar, Mahdi and Ejtemaei Mehr, Shahram and Hassanipour, Mahsa and Mashhadi Akbar Boojar, Masoud and Dehpour, Ahmad Reza
Journal: Iranian Journal of Pharmaceutical Research (2016): 421--433
Authors: Baixauli, Francesc and Acín-Pérez, Rebeca and Villarroya-Beltrí, Carolina and Mazzeo, Carla and Nunez-Andrade, Norman and Gab, undefined and é-Rodriguez, Enrique and Ledesma, Maria Dolores and Blázquez, Alberto and Martin, Miguel Angel and Falcón-Pérez, Juan Manuel and others, undefined
Journal: Cell metabolism (2015): 485--498
Authors: Davis, Warren
Journal: Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids (2014): 168--179
Authors: Xu, Miao and Liu, Ke and Southall, Noel and Marugan, Juan J and Remaley, Alan T and Zheng, Wei
Journal: Analytical and bioanalytical chemistry (2012): 407--414
References
Authors: Urbina P, Flores-Diaz M, Alape-Giron A, Alonso A, Goni FM.
Journal: Chem Phys Lipids (2009): 51
Authors: Tian HP, Qiu TZ, Zhao J, Li LX, Guo J.
Journal: Brain Inj (2009): 1073
Authors: de Santi Ferrara GI, Fern and es-Pedrosa Mde F, Junqueira-de-Azevedo Ide L, Goncalves-de-Andrade RM, Portaro FC, Manzoni-de-Almeida D, Murakami MT, Arni RK, van den Berg CW, Ho PL, Tambourgi DV.
Journal: Toxicon (2009): 743
Authors: Ichi I, Kamikawa C, Nakagawa T, Kobayashi K, Kataoka R, Nagata E, Kitamura Y, Nakazaki C, Matsura T, Kojo S.
Journal: Toxicology (2009): 33
Authors: Sugimori D., undefined
Journal: J Biosci Bioeng (2009): 293
Authors: Ito H, Murakami M, Furuhata A, Gao S, Yoshida K, Sobue S, Hagiwara K, Takagi A, Kojima T, Suzuki M, Banno Y, Tanaka K, Tamiya-Koizumi K, Kyogashima M, Nozawa Y, Murate T.
Journal: Biochim Biophys Acta (2009): 681
Authors: Hiukka A, Stahlman M, Pettersson C, Levin M, Adiels M, Teneberg S, Leinonen ES, Hulten LM, Wiklund O, Oresic M, Olofsson SO, Taskinen MR, Ekroos K, Boren J.
Journal: Diabetes (2009): 2018
Authors: Oda M., undefined
Journal: Yakugaku Zasshi (2009): 1233
Authors: Andersson D, Kotarsky K, Wu J, Agace W, Duan RD.
Journal: Dig Dis Sci (2009): 1440
Authors: Vasil ML, Stonehouse MJ, Vasil AI, Wadsworth SJ, Goldfine H, Bolcome RE, 3rd, Chan J.
Journal: PLoS Pathog (2009): e1000420
Application notes
Design of potent inhibitors of acetylcholinesterase using morin as the starting compound
Acetylcholinesterase Inhibitory Activity of Pigment Echinochrome A
Induction of Neurite Outgrowth in PC12 Cells
Induction of Neuritogenesis in PC12 Cells by a Pulsed Electromagnetic Field
FAQ
How should I reconstitute an NADPH standard?
Will Amplite® Fluorimetric NAD/NADH Ratio Assay Kit *Red Fluorescence* work with NADP/NADPH? Can this kit measure NADP+ and NADPH?
What is the concentration of calcium inside cells?
What assay kits measure NADP/NADPH from cell samples?