Cal-590™ AM
Example protocol
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Prepare a 2 to 5 mM stock solution of Cal-590™ AM in anhydrous DMSO.
Note: When reconstituted in DMSO, Cal-590™ AM is a clear, colorless solution.
PREPARATION OF WORKING SOLUTION
On the day of the experiment, either dissolve Cal-590™ AM in DMSO or thaw an aliquot of the indicator stock solution to room temperature.
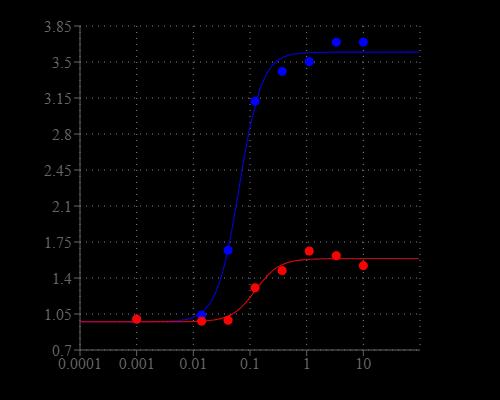
Prepare a 2 to 20 µM Cal-590™ AM working solution in a buffer of your choice (e.g., Hanks and Hepes buffer) with 0.04% Pluronic® F-127. For most cell lines, Cal-590™ AM at a final concentration of 4-5 μM is recommended. The exact concentration of indicators required for cell loading must be determined empirically.
Note: The nonionic detergent Pluronic® F-127 is sometimes used to increase the aqueous solubility of Cal-590™ AM. A variety of Pluronic® F-127 solutions can be purchased from AAT Bioquest.
Note: If your cells contain organic anion-transporters, probenecid (1-2 mM) may be added to the dye working solution (final in well concentration will be 0.5-1 mM) to reduce leakage of the de-esterified indicators. A variety of ReadiUse™ Probenecid products, including water-soluble, sodium salt, and stabilized solutions, can be purchased from AAT Bioquest.
SAMPLE EXPERIMENTAL PROTOCOL
Following is our recommended protocol for loading AM esters into live cells. This protocol only provides a guideline and should be modified according to your specific needs.
- Prepare cells in growth medium overnight.
On the next day, add 1X Cal-590™ AM working solution to your cell plate.
Note: If your compound(s) interfere with the serum, replace the growth medium with fresh HHBS buffer before dye-loading.
Incubate the dye-loaded plate in a cell incubator at 37 °C for 30 to 60 minutes.
Note: Incubating the dye for longer than 2 hours can improve signal intensities in certain cell lines.
- Replace the dye working solution with HHBS or buffer of your choice (containing an anion transporter inhibitor, such as 1 mM probenecid, if applicable) to remove any excess probes.
- Add the stimulant as desired and simultaneously measure fluorescence using either a fluorescence microscope equipped with a TRITC/Cy3 filter set or a fluorescence plate reader containing a programmable liquid handling system such as an FDSS, FLIPR, or FlexStation, at Ex/Em = 540/590 nm cutoff 570 nm.
Calculators
Common stock solution preparation
| 0.1 mg | 0.5 mg | 1 mg | 5 mg | 10 mg | |
| 1 mM | 78.938 µL | 394.692 µL | 789.384 µL | 3.947 mL | 7.894 mL |
| 5 mM | 15.788 µL | 78.938 µL | 157.877 µL | 789.384 µL | 1.579 mL |
| 10 mM | 7.894 µL | 39.469 µL | 78.938 µL | 394.692 µL | 789.384 µL |
Molarity calculator
| Mass (Calculate) | Molecular weight | Volume (Calculate) | Concentration (Calculate) | Moles | ||||
| / | = | x | = |
Spectrum
Product family
| Name | Excitation (nm) | Emission (nm) | Quantum yield |
| Cal-500™ AM | 388 | 482 | 0.481 |
| Cal-520®, AM | 492 | 515 | 0.751 |
| Cal-520ER™ AM | 492 | 515 | - |
| Cal-520FF™, AM | 492 | 515 | 0.751 |
| Cal-520N™, AM | 492 | 515 | 0.751 |
| Cal-630™ AM | 609 | 626 | 0.371 |
| Calbryte™ 590 AM | 581 | 593 | - |
Citations
Authors: Rusakov, Dmitri and Savtchenko, Leonid and Jensen, Thomas and Zheng, Kaiyu
Journal: (2025)
Authors: Zhang, Yunxia and Wu, Qiqian and Bai, Furong and Hu, Yanqin and Xu, Bufang and Tang, Yujie and Wu, Jingwen
Journal: Journal of Ovarian Research (2025): 75
Authors: Spurlock, Brian M and Xie, Yifang and Song, Yiran and Ricketts, Shea N and Hua, James Rock and Chi, Haley R and Nishtala, Meenakshi and Salmenov, Rustem and Liu, Jiandong and Qian, Li
Journal: Cell Reports (2025)
Authors: Wang, Lin
Journal: (2024)
Authors: Bonner, Caroline and Saponaro, Chiara and Imbernon, Monica and Louvet, Isaline and Deligia, Eleonora and Chen, Shiqian and Davies, Iona and Acosta-Montalvo, Ana and Moreno-Lopez, Maria and Wemelle, Eve and others,
Journal: (2024)