Amplite® Fluorimetric NAD/NADH Ratio Assay Kit
Red Fluorescence
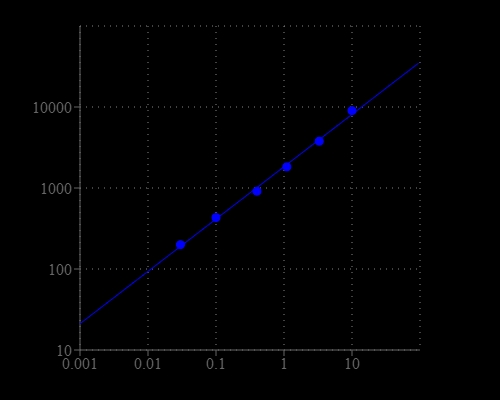
Amplite® Fluorimetric NAD/NADH Ratio Assay Kit enables sensitive fluorescence-based quantification of NAD, NADH, and their ratio in biological samples.
- Sensitive cycling assay: Uses enzyme-coupled cycling reaction to amplify detection of NAD/NADH
- Red fluorescence readout: Minimizes background interference with visible-range detection
- Application versatility: Suitable for metabolism studies, mitochondrial function assays, and redox state analysis
- Comparable alternative: Fluorescence-based substitute for Sigma’s NAD/NADH quantification kits

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 15263 | 250 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 540 nm |
| Emission | 590 nm |
| Cutoff | 570 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 24, 2026
