Amplite® Universal Fluorimetric Protease Activity Assay Kit
Red Fluorescence
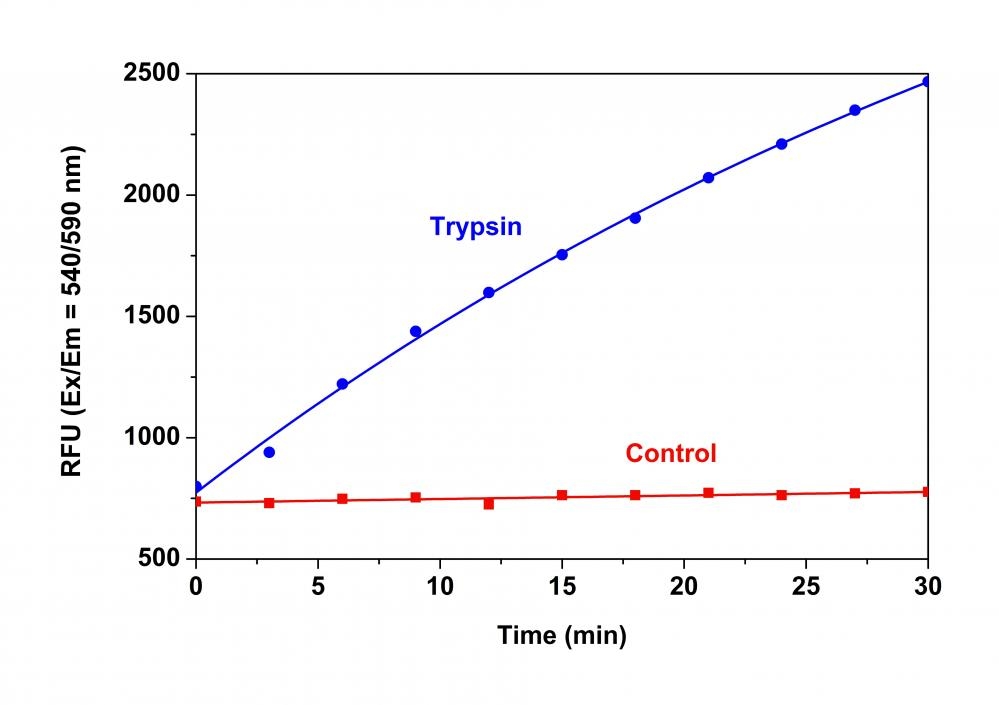
Monitoring of various protease activities has become a routine task for many biological laboratories. Our Amplite® Universal Fluorimetric Protease Activity Assay Kits are an ideal choice for performing routine assays necessary during the isolation of proteases, or for identifying the presence of contaminating proteases in protein samples. The kits use fluorescent casein conjugates that are proven to be a generic substrate for a broad spectrum of proteases. In the intact substrate, casein is heavily labeled with a fluorescent dye, resulting in significant fluorescence quenching. Protease-catalyzed hydrolysis relieves its quenching effect, yielding brightly fluorescent dye-labeled short peptides. The increase in fluorescence intensity is directly proportional to protease activity. The kits provide all the essential components with an optimized "mix & read" protocol that can be easily automated to HTS instruments.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 13501 | 500 Tests | Price |
Spectral properties
| Correction factor (260 nm) | 0.32 |
| Correction factor (280 nm) | 0.178 |
| Extinction coefficient (cm -1 M -1) | 90000 |
| Excitation (nm) | 552 |
| Emission (nm) | 578 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 540 nm |
| Emission | 590 nm |
| Cutoff | 570 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 4, 2026

