Covidyte™ EN450
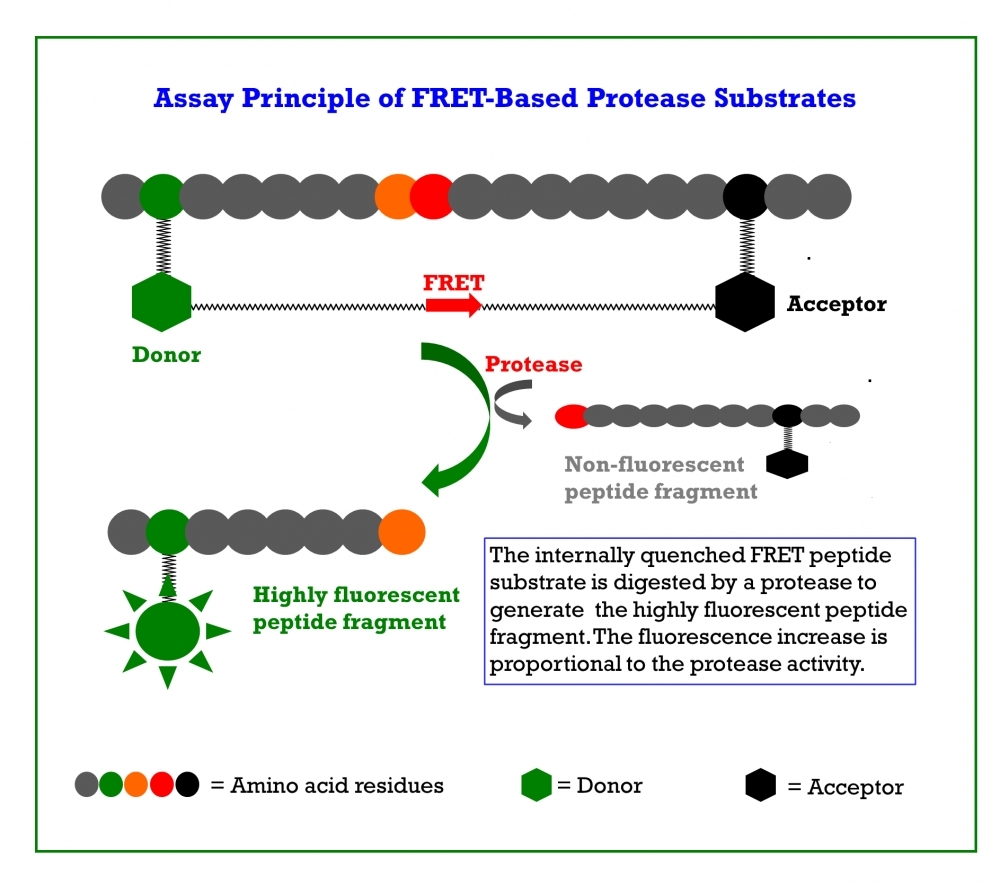
Coronaviruses (CoVs) can infect humans and multiple species of animals, causing a wide spectrum of diseases. In late 2019, a novel coronavirus, termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was determined as a cause for several cases of respiratory disease (Covid-19). Even though most infected patients only suffer from mild symptoms such as fever and cough associated with a good prognosis, the disease can progress into fatal cases of pneumonia and acute respiratory failure, especially in older males with comorbidities. The virus rapidly spread worldwide. As of May 31st, 2022, more than 6.2 million people have died from coronavirus worldwide, and ~530 million cases have been reported. Currently, there are not any specific and effective options available for treating Covid-19. At present the clinical treatment of Covid-19 is mainly symptomatic combined with repurposing of already marketed antiviral drugs such as Remdesivir and antibiotics to treat secondary infections. There is an extremely urgent need for the development of specific antiviral therapeutics and vaccines against SARS-CoV-2. The coronavirus main protease, which plays a pivotal role in viral gene expression and replication through the proteolytic processing of replicase polyproteins, is an attractive target for anti-CoV drug design. The inhibition of viral proteases necessary for proteolytic processing of polyproteins has been a successful strategy in the treatment of human immunodeficiency virus (HIV) and hepatitis C respectively, proving the potential of protease inhibitors for the treatment of viral infections. Similarly, the main protease of SARS-CoV-2 is thought to be essential for viral replication and, therefore, is regarded as promising target for antiviral therapy of Covid-19. Covidyte™ EN450 is a peptide substrate containing 14 amino acid sequence (KTSAVLQSGFRKME) that can be cleaved by coronavirus proteases. The dark-FRET peptide contains Dabcyl (quencher) and Edans (donor) on the N-and C-terminals respectively where the fluorescence of Edans is effectively quenched by Dabcyl when the peptide is intact. When the peptide is hydrolyzed by coronavirus proteases, the Edans fragment generates significantly enhanced fluorescence since its fluorescence is no longer quenched by Dabcyl. The activity of coronavirus proteases can be effectively monitored by the fluorescence intensity of Edans. Covidyte™ EN450 is a convenient tool for screening inhibitors of coronavirus proteases.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 13535 | 100 tests | Price | |
| 13536 | 1000 tests | Price |
Physical properties
| Molecular weight | 2584.98 |
| Solvent | DMSO |
Spectral properties
| Correction factor (280 nm) | 0.107 |
| Extinction coefficient (cm -1 M -1) | 5900 |
| Excitation (nm) | 336 |
| Emission (nm) | 455 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 350 nm |
| Emission | 460 nm |
| Cutoff | 420 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 15, 2026

