Amplite® Colorimetric Sphingomyelinase Assay Kit
Blue Color
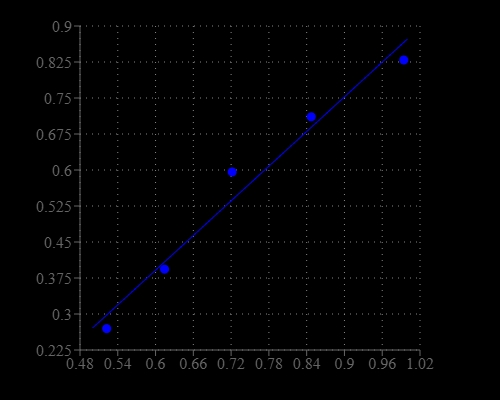
Sphingomyelinase (SMase) is an enzyme that is responsible for cleaving sphingomyelin (SM) to phosphocholine and ceramide. Activation of SMases in cells plays an important role in the cellular responses. Five types of sphingomyelinase (SMase) have been identified based on their cation dependence and pH optima of action. They are lysosomal acid SMase, secreted zinc-dependent acid SMase, magnesium-dependent neutral SMase, magnesium-independent neutral SMase, and alkaline SMase. Among the five types, the lysosomal acidic SMase and the magnesium-dependent neutral SMase are considered major candidates for the production of ceramide in the cellular response to stress. Our Amplite® Colorimetric Sphingomyelinase Assay Kit provides a sensitive method for detecting neutral SMase activity or screening its inhibitors. The kit uses Amplite® UltraBlue as a colorimetric probe to indirectly quantify the phosphocholine produced from the hydrolysis of sphingomyelin (SM) by sphingomyelinase (SMase). It can be used for measuring the SMase activity in blood, cell extracts or other solutions. The absorbance of light at 655 nm is proportional to the formation of phosphocholine, therefore to the SMase activity. The kit is an optimized "mix and read" assay that is compatible with HTS liquid handling instruments.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 13620 | 200 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Absorbance microplate reader | |
| Absorbance | 655 nm |
| Recommended plate | Clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 2, 2026
