Amplite® Fluorimetric Glucose Quantitation Kit
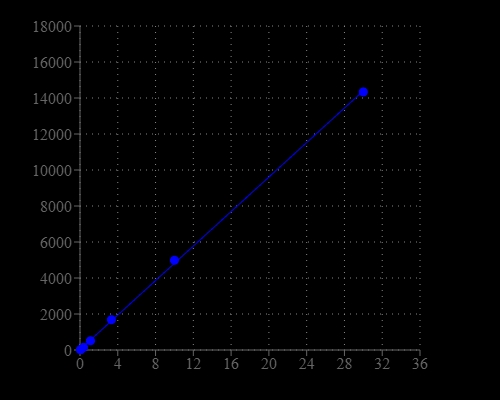
Glucose, a monosaccharide, is the most important carbohydrate in biology. It is a source of energy and metabolic intermediate for cell growth. Glucose is one of the main products of photosynthesis and starts cellular respiration in both prokaryotes and eukaryotes. Glucose level is a key diagnostic parameter for many metabolic disorders. This glucose assay kit provides a quick and sensitive method for the measurement of glucose in various biological samples (e.g., serum, plasma, body fluid, food, growth medium, etc.). The kit uses our Amplite®Red substrate that making the kit recordable in a dual more, either fluorimetric or colorimetric readout. The kit provides all the essential components with an optimized assay protocol. The assay is robust, and can be readily adapted for high-throughput assays in a wide variety of applications that require the measurement of glucose. For example, the assay might be suitable for monitoring glucose level during fermentation and glucose feeding in protein expression processes. It might also be used for monitoring glucose transporters.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 40005 | 500 Tests | Price |
Spectral properties
| Excitation (nm) | 571 |
| Emission (nm) | 584 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 540 nm |
| Emission | 590 nm |
| Cutoff | 570 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 15, 2026

