Amplite® Fluorimetric Proteasome 20S Activity Assay Kit
Green Fluorescence
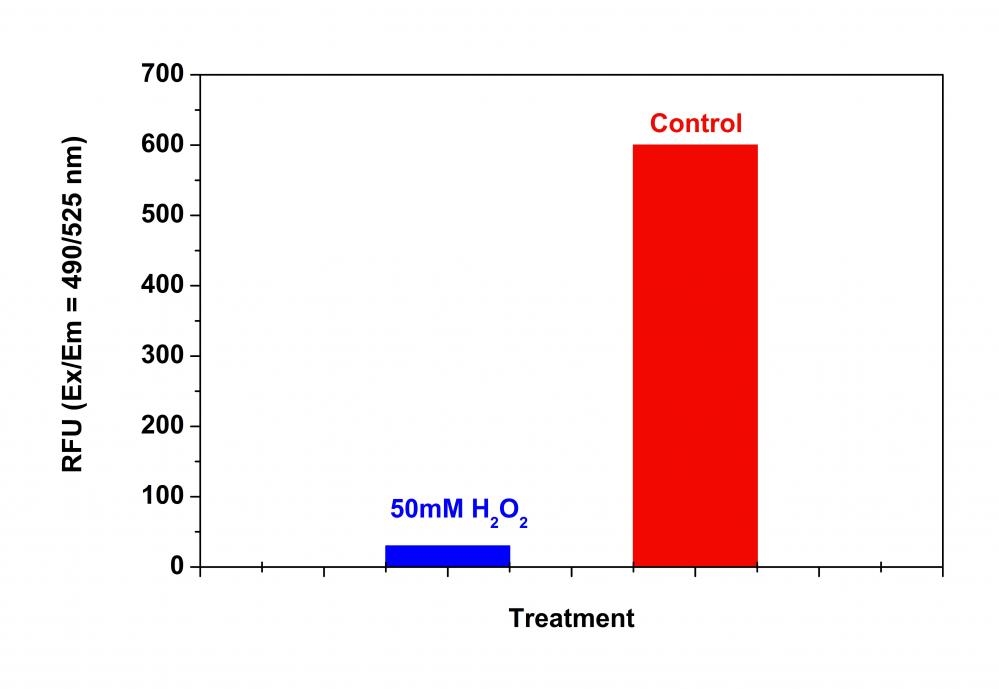
Our Amplite® Fluorometric 20S Proteasome Assay Kit is a homogeneous fluorescent assay that measures the chymotrypsin-like protease activity associated with the proteasome complex either in cultured cells or cell lysates. This kit uses LLVY-R110 as a fluorogenic indicator for proteasome activity. Cleavage of LLVY-R110 by proteasome generates the strongly green fluorescent R110 that is monitored fluorimetrically at 520-530 nm with excitation of 480-500 nm. The kit provides all the essential components with an optimized assay protocol. The assay is robust, and can be readily adapted for high-throughput assays for evaluation of proteasome activity or inhibitor screening in cultured cells or in solution. The assay can be performed in a convenient 96-well and 384-well fluorescence microtiter-plate format. The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The most common form of the proteasome in this pathway is the 26S proteasome, an ATP-dependent proteolytic complex, which contains one 20S (700-kDa) core particle structure and two 19S (700-kDa) regulatory caps. The 20S core contains three major proteolytic activities including chymotrypsin-like, trypsin-like and caspase-like. It is responsible for the breakdown of key proteins involved with apoptosis, DNA repair, endocytosis, and cell cycle control.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 13456 | 100 tests | Price |
Spectral properties
| Extinction coefficient (cm -1 M -1) | 80000 |
| Excitation (nm) | 500 |
| Emission (nm) | 522 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Solid black |
| Instrument specification(s) | Top read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 5, 2026

