Amplite® Luciferase Reporter Gene Assay Kit
Maximized Luminescence
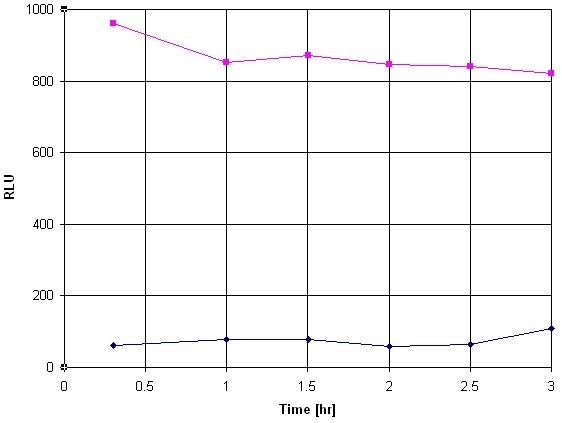
Common reporter genes include beta-galactosidase, beta-glucuronidase and luciferase. The advantages of a luciferase assay are the high sensitivity, the absence of luciferase activity inside most of the cell types, the wide dynamic range, rapidity and low cost. The most versatile and common reporter gene is the luciferase of the North American firefly Photinus pyralis. The protein requires no posttranslational modification for enzyme activity. It is not even toxic in high concentration (in vivo) and can be used in pro- and eukaryotic cells. The firefly luciferase catalyzes the bioluminescent oxidation of luciferin in the presence of ATP, magnesium and oxygen. This Amplite® Luciferase Reporter Gene Assay Kit uses a proprietary luminogenic formulation to quantify luciferase activity in live cells and cell extracts. Our formulation generates a luminescent product that gives strong luminescence upon interaction with luciferase. The kit provides all the essential components with our optimized 'mix and read' assay protocol that is compatible with HTS liquid handling instruments. It has extremely high sensitivity, and can be used for the assays that require demanding sensitivity.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 12518 | 1 plate | Price | |
| 12519 | 10 plates | Price | |
| 12520 | 100 plates | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 41105331 |
Instrument settings
| Luminescence microplate reader | |
| Recommended plate | Solid white |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 30, 2026
