Amplite® Universal Fluorimetric Kinase Assay Kit
Red Fluorescence
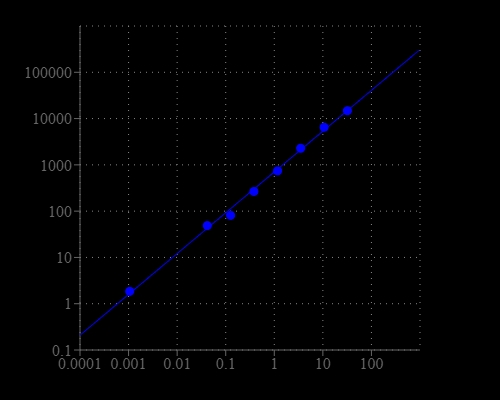
Most of commercial protein kinase assay kits are either based on monitoring of phosphopeptide formation or ATP deletion. For the kinase assay kits that are based on detection of phosphopeptides one has to spend time and efforts to identify an optimized peptide substrate while the ATP depletion method suffers various interferences due to the use of luciferase that are inhibited or activated by various biological compounds. The Amplite® Universal Kinase Assay Kit is based on the monitoring of ADP formation, which is directly proportional to enzyme phoshphotransferase activity and is measured fluorimetricaly. This kit provides a fast, simple, and homogeneous assay for measure kinases activities. The characteristics of its high sensitivity (<0.2 uM ADP), broad ATP tolerance (1-300 uM), non-antibody based, non-radioactive and no-wash method to detect the amount of ADP produced as a result of enzyme activity make it an ideal kit for determining kinase Michaelis-Menten kinetics and for screening and identifying kinase inhibitors. The assay can be performed in a convenient 96-well or 384-well microtiter-plate format and easily adapted to automation with no separation steps required.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 31001 | 250 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 540 nm |
| Emission | 590 nm |
| Cutoff | 570 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026
