Cell Meter™ Autophagy Assay Kit
Green Fluorescence
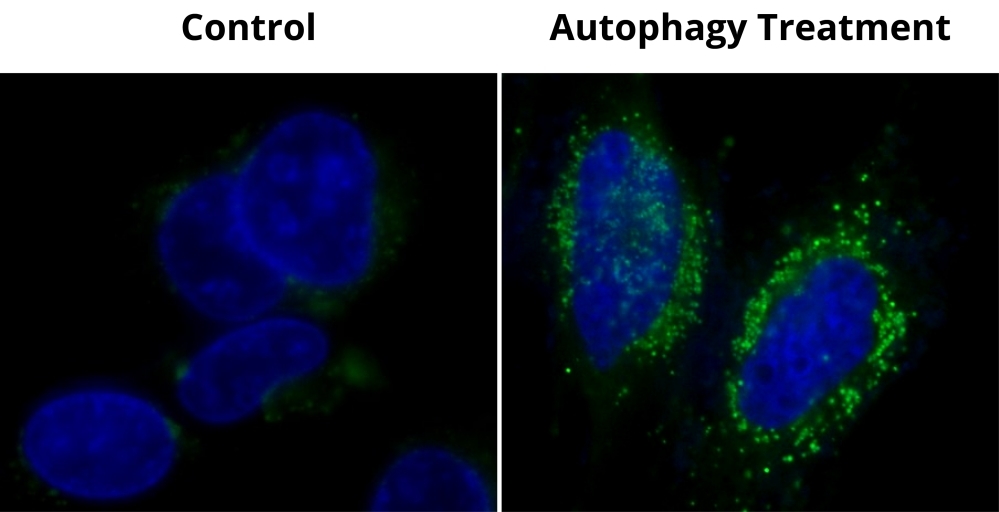
This Cell Meter™ Autophagy Kit employs Autophagy Green™ as a specific autophagosome marker to analyze the activity of autophagy. The assay is optimized for direct detection of autophagy in both detached and attached cells. The kit provides all the essential components for the assay protocol. Cell Meter™ Autophagy Kit is suitable for fluorescence microscope, fluorescence microplate reader and flow cytometer. Autophagy Green™ has a large Stokes shift. Autophagy is an evolutionarily conserved degradation process that targets long-lived proteins, organelles, and other cytoplasmic components for degradation via the lysosomal pathway. The autophagy pathway is complementary to the action of the ubiquitin-proteasome pathway which typically degrades short-lived proteins. Activation of the autophagy pathway is required for multiple cellular roles, including survival during starvation, the clearance of intracellular components, development, and immunity. In the absence of stress, autophagy serves a house-keeping function, removing damaged organelles and cellular components preventing cytotoxic effects. Decreases and defects in autophagy have been implicated in multiple diseases, for example Huntingtons, Alzheimers, and Parkinsons. In terms of cancer development, autophagy seems to play multiple roles. Decreased or absent expression of certain autophagy proteins, such as Beclin-1 and Bif-1, increases tumor susceptibility in mice while the overexpression of these proteins can repress cancer cell growth. However, autophagy is critical for the survival of cancer cells within the nutrient poor and hypoxic core of solid tumors.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 23002 | 200 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm laser |
| Emission | 530/30 nm filter |
| Instrument specification(s) | FITC channel |
| Fluorescence microscope | |
| Excitation | FITC filter |
| Emission | FITC filter |
| Recommended plate | Black wall/clear bottom |
| Fluorescence microplate reader | |
| Excitation | 485 nm |
| Emission | 530 nm |
| Cutoff | 515 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 20, 2026
