Cell Meter™ Fluorimetric Live Cell Cycle Assay Kit
Green Fluorescence Optimized for Flow Cytometry
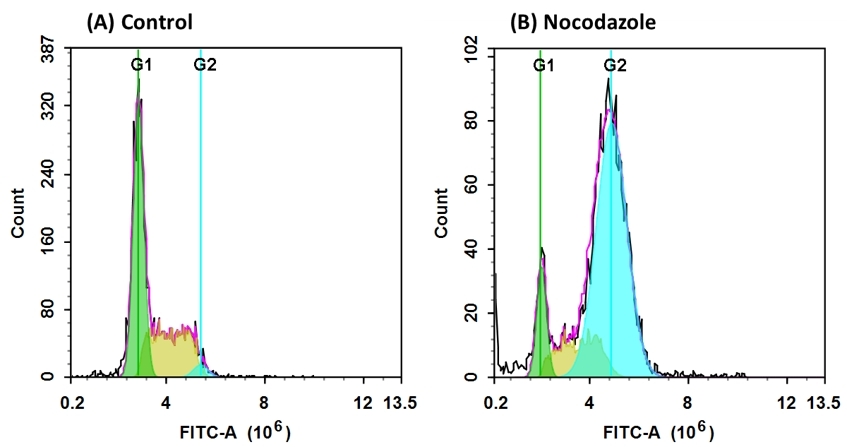
Our Cell Meter™ assay kits are a set of tools for monitoring cell viability and proliferation. There are a variety of parameters that can be used for monitoring cell viability and proliferation. In normal cells, DNA density changes depending on whether the cell is growing, dividing, resting, or performing its ordinary functions. The progression of the cell cycle is controlled by a complex interplay among various cell cycle regulators. These regulators activate transcription factors, which bind to DNA and turn on or off the production of proteins that result in cell division. Any misstep in this regulatory cascade causes abnormal cell proliferation which underlies many pathological conditions, such as tumor formation. Potential applications for live-cell studies are in the determination of cellular DNA content and cell cycle distribution for the detection of variations in growth patterns, for monitoring apoptosis, and for evaluating tumor cell behavior and suppressor gene mechanisms. This particular kit is designed to monitor cell cycle progression and proliferation using our proprietary Nuclear Green™ LCS1 in live, permeabilized and fixed cells. The percentage of cells in a given sample that are in G0/G1, S and G2/M phases, as well as the cells in the sub-G1 phase prior to apoptosis can be determined by flow cytometry. Cells stained with Nuclear Green™ LCS1 can be monitored with a flow cytometer (FL1 channel).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22841 | 100 Tests | Price |
Spectral properties
| Excitation (nm) | 503 |
| Emission (nm) | 527 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm laser |
| Emission | 530/30 nm filter |
| Instrument specification(s) | FITC channel |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 15, 2026

