Cell Meter™ Fluorimetric Live Cell Cycle Assay Kit
Optimized for 405 nm Violet Laser Excitation
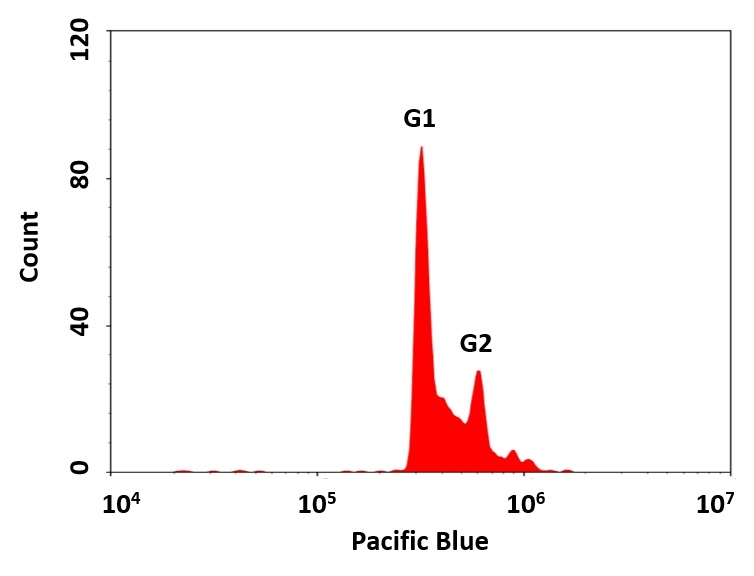
The cell cycle has four sequential phases: G0/G1, S, G2, and M. During a cell's passage through cell cycle, its DNA is duplicated in S (synthesis) phase and distributed equally between two daughter cells in M (mitosis) phase. These two phases are separated by two gap phases: G0/G1 and G2. The two gap phases provide time for the cell to grow and double the mass of their proteins and organelles. They are also used by the cells to monitor internal and external conditions before proceeding with the next phase of cell cycle. The cell's passage through cell cycle is controlled by a host of different regulatory proteins. This particular kit is designed to monitor cell cycle progression and proliferation by using our proprietary Nuclear Violet™ in live cells. The percentage of cells in a given sample that are in G0/G1, S and G2/M phases, as well as the cells in the sub-G1 phase prior to apoptosis can be determined by flow cytometry. Cells stained with Nuclear Violet™ can be monitored with a flow cytometer (Pacific Blue® channel).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22845 | 100 Tests | Price |
Spectral properties
| Excitation (nm) | 401 |
| Emission (nm) | 460 |
Storage, safety and handling
| Intended use | Research Use Only (RUO) |
Instrument settings
| Flow cytometer | |
| Excitation | 405 nm laser |
| Emission | 450/40 nm filter |
| Instrument specification(s) | Pacific Blue channel |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 27, 2026

