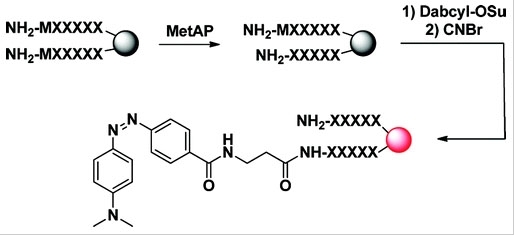
DABCYL succinimidyl ester
4-((4-(Dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester; CAS 146998-31-4
DABCYL, SE is the amino-reactive form of DABCYL, and widely used to prepare a variety of FRET-based probes that contain DABCYL. DABCYL is one of the most popular acceptors for developing FRET-based nucleic acid probes and protease substrates.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 2004 | 1 g | Price | |
| 2005 | 5 g | Price |
Physical properties
| Molecular weight | 366.37 |
| Solvent | DMSO |
Spectral properties
| Absorbance (nm) | 454 |
| Correction factor (280 nm) | 0.516 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12352200 |
| CAS | 146998-31-4 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026

