Gelite™ Green Nucleic Acid Gel Staining Kit
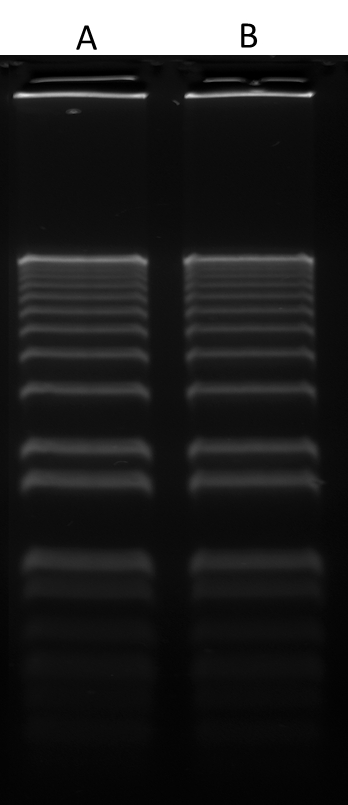
Gelite™ Green is a sensitive fluorescent nucleic acid gel stain for detecting nucleic acids in agarose and polyacrylamide gels. Gelite™ Green stain exhibits exceptional affinity for DNA and a large fluorescence enhancement upon binding to DNA, at least an order of magnitude greater than that of ethidium bromide when detected by photography. With a standard 300 nm UV transilluminator and photographic detection, as little as 60 pg dsDNA per band can be detected with Gelite™ Green stain. Gelite™ Green nucleic acid gel stain is nearly two orders of magnitude more sensitive than ethidium bromide for staining oligonucleotides in gels. Our Gelite™ Green Nucleic Acid Gel Staining Gel Kit includes our Gelite™ Green nucleic acid stain with an optimized and robust protocol. It provides a convenient solution for staining nucleic acid samples in gels.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17589 | 1 Kit | Price |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| UNSPSC | 41116134 |
Instrument settings
| Transilluminator | |
| Excitation | 254 nm or 300 nm |
| Emission | Long path green filter (ex. SYBR or GelStar) |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 30, 2026
