Helixyte™ iFluor® 750 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA*
Example protocol
AT A GLANCE
Combine DNA with the Helixyte™ iFluor® 750 Nucleic Acid Labeling Dye stock solution.
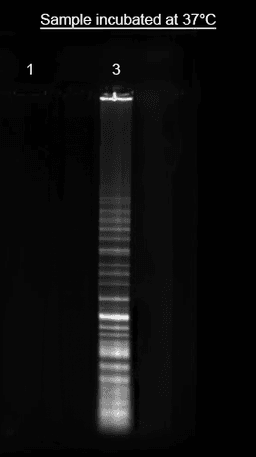
Incubate for 1 hour at 37°C.
Purify the conjugate as required for downstream applications.
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Before opening the vial, thaw Helixyte™ iFluor® nucleic acid labeling dye at room temperature. Briefly centrifuge to collect the dried pellet.
Add 70 μL of DMSO to the Helixyte™ iFluor® 750 Nucleic Acid Labeling Dye vial to prepare a 10 mM stock solution.
Note: It is recommended to divide any unused stock solution into single-use aliquots. Store the aliquots at ≤-20 ºC and protect them from light. Avoid repeated freeze-thaw cycles.
SAMPLE EXPERIMENTAL PROTOCOL
Prepare the labeling reaction according to the specifications in table 1 below.
Table 1. Standard Nucleic Acid Labeling Reaction.
Components Volume added to reaction Final Concentration DNA (1 mg/mL) 2 to 5 µL 2 to 5 µg Helixyte™ iFluor® 750 Nucleic acid Labeling Dye stock solution 1 µL 100 µM TE Buffer (pH 8 to 8.5) Add sufficient buffer to adjust the volume to 100 µL Note: This DNA:Dye ratio results in labeling efficiencies that are appropriate for most applications. The amount of Helixyte™ iFluor® 750 Nucleic Acid Labeling Dye or the reaction incubation time can be adjusted to modify the labeling density as per the application requirements. The DNA-to-dye ratio must be optimized to achieve a higher labeling ratio.
Incubate the reaction at 37℃ for 1 hour, protected from light.
Note: After 30 minutes of incubation, briefly centrifuge the reaction to minimize the effects of evaporation and maintain the appropriate concentration of the reaction components.
Note: Alternatively, the reaction can be incubated at room temperature for 2 hours. For the best labeling condition, we recommend incubating at 37℃.
After incubation, the labeling mix can be purified to remove any free labeling dye. Refer to the “Purification of labeling mix with alcohol precipitation” section below for instructions.
Add 0.1 volume (10 µL) of 5M sodium chloride and 2 - 2.5 volumes of ice-cold 100% ethanol (250 µL) to the reaction. Mix well and place at ≤ -20°C for at least 30 minutes.
Centrifuge at full speed (>14,000 x g) in a refrigerated micro centrifuge for 15-30 minutes to pellet the labeled nucleic acid. Once pelleted, carefully remove the ethanol with a micropipette. Do not disturb the pellet.
Note: Small nucleic acid quantities can be difficult to visualize. Mark and orient the precipitate-containing tubes in the microfuge such that the pellet will form in a predetermined place.
Wash the pellet once with 500 μL of room temperature 70% ethanol. Centrifuge at full speed for an additional 15-30 minutes.
Remove all traces of ethanol with a micropipette. DO NOT allow the sample to dry longer than 5 minutes as the pellet may become difficult to resuspend.
Resuspend the labeled DNA with ~ 30 µL sterile water.
Store the purified, labeled nucleic acid for long-term storage or put on ice for immediate use.
Spectrum
Product family
| Name | Excitation (nm) | Emission (nm) | Extinction coefficient (cm -1 M -1) | Quantum yield | Correction Factor (260 nm) | Correction Factor (280 nm) |
| Helixyte™ iFluor® 350 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 345 | 450 | 200001 | 0.951 | 0.83 | 0.23 |
| Helixyte™ iFluor® 488 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 491 | 516 | 750001 | 0.91 | 0.21 | 0.11 |
| Helixyte™ iFluor® 555 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 557 | 570 | 1000001 | 0.641 | 0.23 | 0.14 |
| Helixyte™ iFluor® 594 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 587 | 603 | 2000001 | 0.531 | 0.05 | 0.04 |
| Helixyte™ iFluor® 647 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 656 | 670 | 2500001 | 0.251 | 0.03 | 0.03 |
References
Authors: Chovelon, Benoît and Peyrin, Eric and Ragot, Mailys and Salem, Nassim and Nguyen, Truong Giang and Auvray, Benjamin and Henry, Mickael and Petrillo, Mel-Alexandre and Fiore, Emmanuelle and Bessy, Quentin and Faure, Patrice and Ravelet, Corinne
Journal: Analytica chimica acta (2023): 340840
Authors: Jin, Xin and Liu, Yingzhu and Alkhamis, Obtin and Canoura, Juan and Bacon, Adara and Xu, Ruyi and Fu, Fengfu and Xiao, Yi
Journal: Analytical chemistry (2022): 10082-10090
Authors: Billet, Blandine and Chovelon, Benoit and Fiore, Emmanuelle and Faure, Patrice and Ravelet, Corinne and Peyrin, Eric
Journal: Biosensors & bioelectronics (2022): 114091
Authors: Koshel, Brooke and Birdsall, Robert and Chen, Weibin
Journal: Journal of chromatography. B, Analytical technologies in the biomedical and life sciences (2020): 121906
Authors: Botequim, David and Silva, Inês I R and Serra, Sofia G and Melo, Eduardo P and Prazeres, Duarte M F and Costa, Sílvia M B and Paulo, Pedro M R
Journal: Nanoscale (2020): 6334-6345