iFluor® 555 maleimide
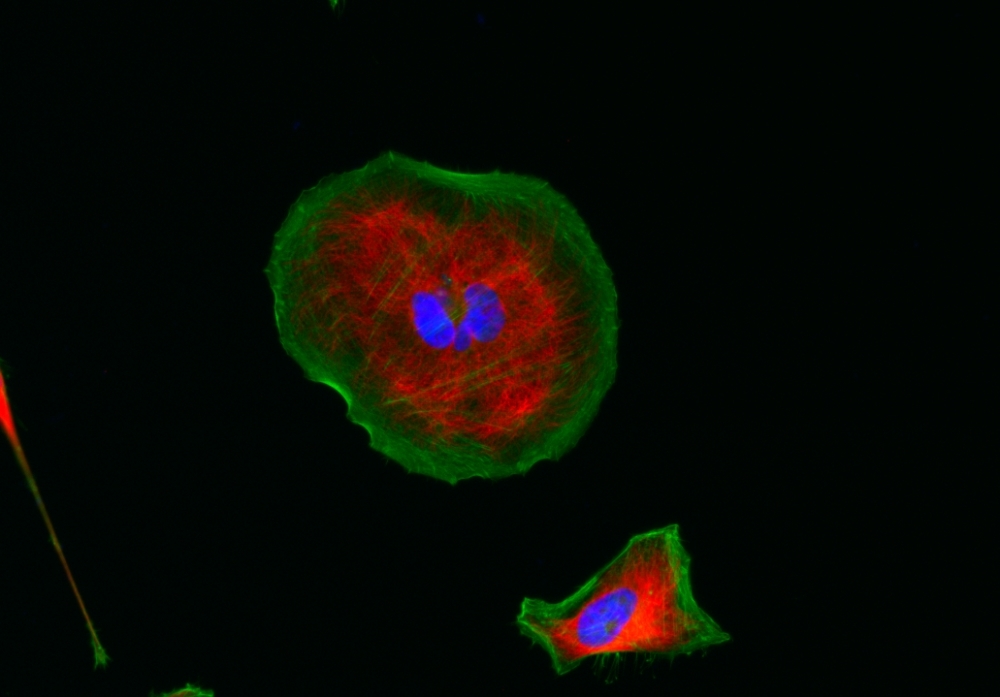
AAT Bioquest's iFluor® dyes are optimized for labeling proteins, particularly antibodies. These dyes are bright, photostable, and have minimal quenching on proteins. They can be well excited by the major laser lines of fluorescence instruments (e.g., 350, 405, 488, 555, and 633 nm). iFluor® 555 dyes have fluorescence excitation and emission maxima of ~557 nm and ~570 nm respectively. The iFluor® 555 family has spectral properties essentially identical to those of Cy3® (Cy3® is the trademark of GE Healthcare). Compared to Cy3 probes, the iFluor® 555 family has much stronger fluorescence and higher photostability. Their fluorescence is pH-independent from pH 3 to 11. These spectral characteristics make this new dye family a superior alternative to Cy3®. iFluor® 555 family has become an excellent replacement for Cy3® and Alexa Fluor® 555 labeling dye (Cy3® and Alexa Fluor® are the trademarks of Invitrogen and GE Health Care). iFluor® 555 maleimide is reasonably stable and shows good reactivity and selectivity with the thiol group.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1063 | 1 mg | Price |
Physical properties
| Molecular weight | 947.93 |
| Solvent | DMSO |
Spectral properties
| Correction factor (260 nm) | 0.23 |
| Correction factor (280 nm) | 0.14 |
| Extinction coefficient (cm -1 M -1) | 100000 1 |
| Excitation (nm) | 557 |
| Emission (nm) | 570 |
| Quantum yield | 0.64 1 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 5, 2026

