MitoLite™ Green FM
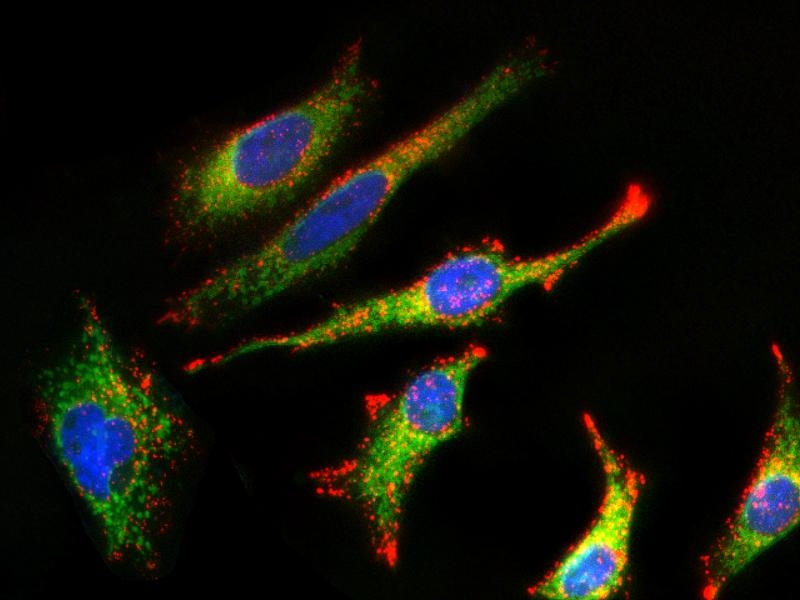
MitoLite™ Green FM is the same molecule to the MitoTracker Green FM (M7514, ThermoFisher). It is green-fluorescent mitochondrial stain. Unlike other MitoLite probes, MitoLite™ Green FM appears to localize to mitochondria, much less depending on mitochondrial membrane potential. The dye stains live cells, but it is not well-retained after aldehyde fixation.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22695 | 10x50 ug | Price |
Physical properties
| Molecular weight | 671.88 |
| Solvent | DMSO |
Spectral properties
| Excitation (nm) | 508 |
| Emission (nm) | 528 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12352200 |
| CAS | 201860-17-5 |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm laser |
| Emission | 530/30 nm filter |
| Instrument specification(s) | FITC channel |
| Fluorescence microscope | |
| Excitation | FITC filter set |
| Emission | FITC filter set |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 18, 2026

