MycoLight™ Fluorimetric CTC Live Bacteria Quantification Kit
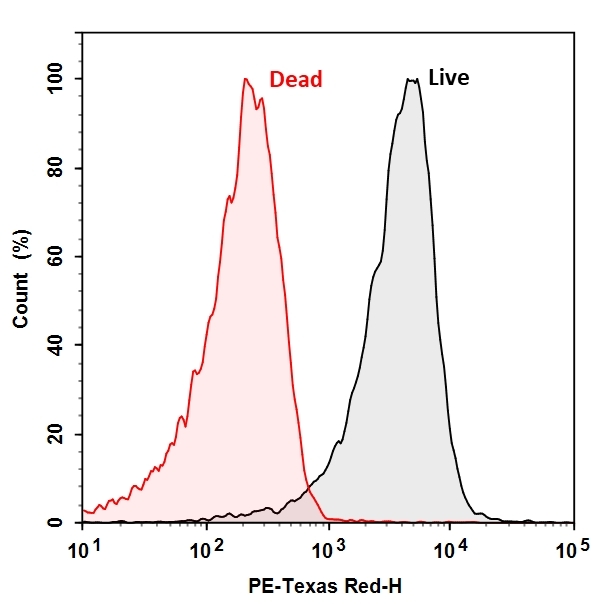
The CTC Flow Cytometric Live Bacteria Assay Kit provides an easy and convenient method for evaluating bacterial health and vitality as a function of the respiratory activity. CTC itself is non-fluorescent, once reduced by the electron transfer system of viable bacterial cell surface, red and insoluble fluorescent CTC formazan is formed and can be detected with a flow cytometer. Dead bacteria that are not respiring or unhealthy bacteria that respire at a lower rate will produce none or less red fluorescent with CTC staining, thus providing a semi-quantitative measure of health status of bacteria population.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22405 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm laser |
| Emission | 610/20 nm filter |
| Instrument specification(s) | PE-Texas Red channel |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 20, 2026
