MycoLight™ Rapid Fluorescence Bacterial Gram Stain Kit
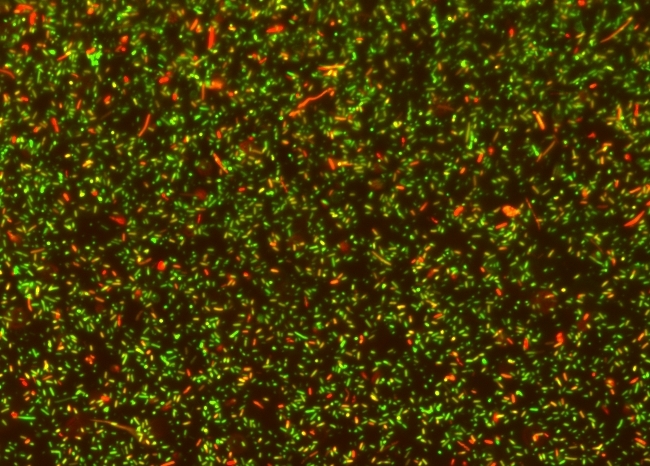
The MycoLight™ Rapid Fluorescence Bacterial Gram Stain Kit provides an easy and convenient way for determination of gram sign in live bacteria. Gram staining is a commonly used method in both clinical and research settings to taxonomically classify bacterial species into two large groups. Unfortunately, the traditional gram staining method is tedious and involves bacterial fixation which can be a significant drawback if the bacteria are to be characterized further. The MycoLight™ Rapid Fluorescence Bacterial Gram Stain Kit provides a one-step gram staining assay for live bacteria that overcomes the problems inherent in the traditional gram staining assays. The MycoLight™ Rapid Fluorescence Bacterial Gram Stain Kit utilizes two DNA dyes MycoLight™ Green and MycoLight™ Red with differential ability to stain gram positive and negative bacteria. MycoLight™ Green stains both gram positive and negative bacteria while MycoLight™ Red preferentially labels gram positive bacteria. The excitation/emission maxima for these two dyes are about 484/504 nm for MycoLight™ Green and 650/669 nm for MycoLight™ Red. Thus, when a mixture of gram positive and gram negative bacteria is stained with the dyes, gram positive bacteria will fluoresce red and gram negative bacteria will fluoresce green. The gram positive and negative staining can be monitored fluorimeterically with Cy5 and FITC filter set respectively.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22413 | 100 Tests | Price |
Spectral properties
| Excitation (nm) | 482 |
| Emission (nm) | 512 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 488/650 nm |
| Emission | 530/669 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | FITC/Cy5 filters |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 19, 2026

