Nuclear Green™ DCS1
5 mM DMSO Solution
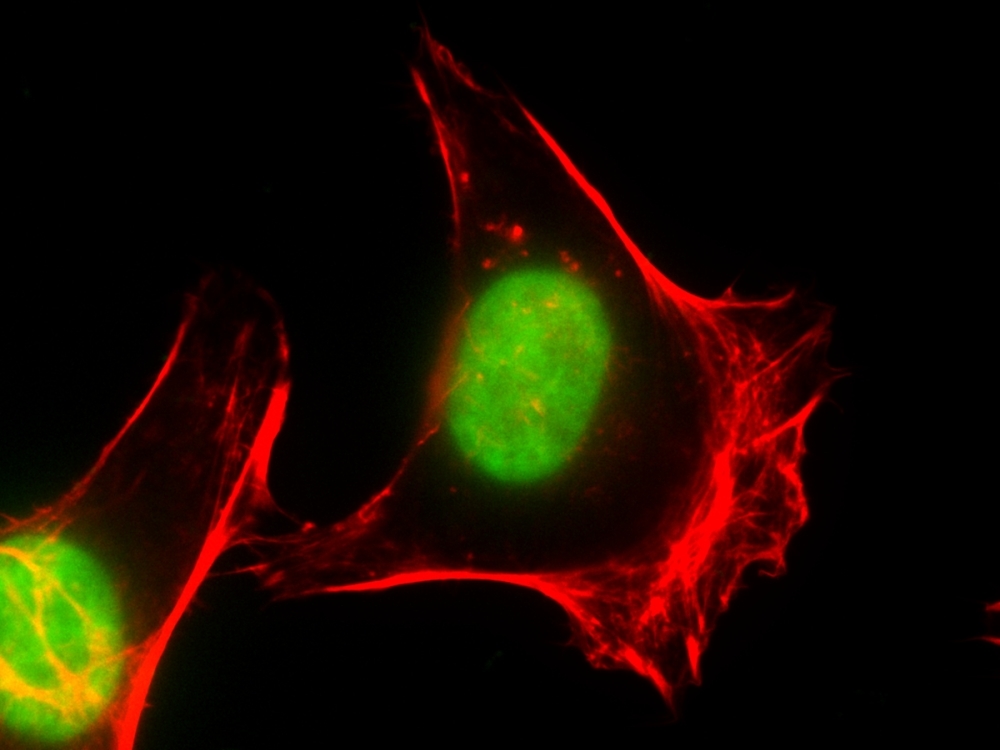
Our Nuclear Green™ DCS1 is a fluorogenic, DNA-selective and cell-impermeant dye for analyzing DNA content in dead, fixed or apoptotic cells. The Nuclear Green™ DCS1 has its green fluorescence significantly enhanced upon binding to DNA. It can be used in fluorescence imaging, microplate and flow cytometry applications. This DNA-binding dye might be used for multicolor analysis of dead, fixed or apoptotic cells with proper filter sets.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17550 | 0.5 ml | Price |
Physical properties
| Solvent | DMSO |
Spectral properties
| Excitation (nm) | 503 |
| Emission (nm) | 527 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 41116134 |
Instrument settings
| Fluorescence microscope | |
| Excitation | FITC filter set |
| Emission | FITC filter set |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026

