PhosphoWorks™ Colorimetric ATP Assay Kit
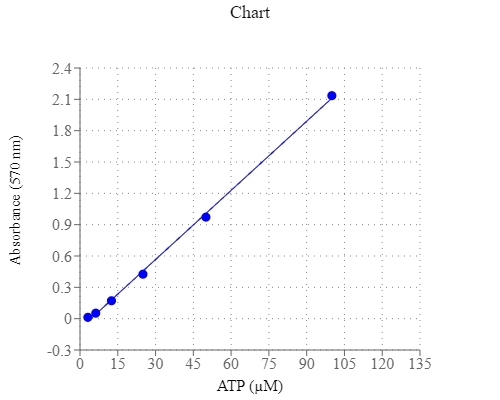
Adenosine triphosphate (ATP) plays a fundamental role in cellular energetics, metabolic regulation and cellular signaling. It is referred as the "molecular unit of currency" of intracellular energy transfer to drive many processes and chemical synthesis in living cells. ATP also serves as a signaling molecule for cell communication and plays an important role in DNA and RNA synthesis. AAT Bioquest offers a variety of bioluminescence assay kits to determine nanomolar (nM) range of ATP with recombinant firefly luciferase (Cat# 21610 & 21609). These kits require luminescence plate readers, are frequently used for cell viability or cytotoxicity assays. PhosphoWorks™ Colorimetric ATP Assay Kit is based on a serial ATP-induced enzyme coupled reactions to produce hydrogen peroxide, which is spectrophotometrically quantified with our Amplite® Red Substrate at OD 570 nm. The assay can detect ~3 µM of ATP in a 100 µL reaction volume with minimal interference from ADP and AMP. It provides a robust, simple and convenient assay for measuring ATP levels in biological samples. The PhosphoWorks™ Colorimetric ATP Assay is complementary to our luciferase-based ATP assay kits.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 21617 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Absorbance microplate reader | |
| Absorbance | 570 nm |
| Recommended plate | Clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 6, 2026
