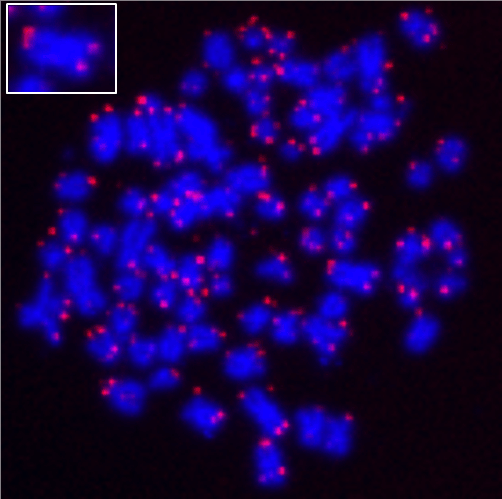
Fluorescence in situ hybridization (FISH) technology is an effective tool for detecting specific nucleic acid targets in a biological specimen. Detection of a nucleic acid target in situ is achieved through the hybridization of a fluorescent dye-labeled nucleic acid probe of complementary sequence to the specimen. The Readilink™ iFluor® 647 FISH fluorescence imaging kit is a convenient tool for labeling a target DNA using an iFluor® 647 labeled FISH probe via in situ hybridization. The kit provides Taq DNA polymerase enzyme which incorporates iFluor® 647-dUTPs in the target DNA through Polymerase Chain Reaction (PCR). Our proprietary iFluor® dyes are brighter and more photostable than traditional fluorescent labels, providing the desired resolution and signal.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17316 | 25 Tests | Price |
| Solvent | Water |
| Correction factor (260 nm) | 0.03 |
| Correction factor (280 nm) | 0.03 |
| Correction factor (656 nm) | 0.0793 |
| Extinction coefficient (cm -1 M -1) | 250000 1 |
| Excitation (nm) | 656 |
| Emission (nm) | 670 |
| Quantum yield | 0.25 1 |
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| Thermal Cycler | |
| Recommended plate | PCR Microplate |
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |

