ReadiLink™ Rapid iFluor® 680 Antibody Labeling Kit
Microscale Optimized for Labeling 50 μg Antibody Per Reaction
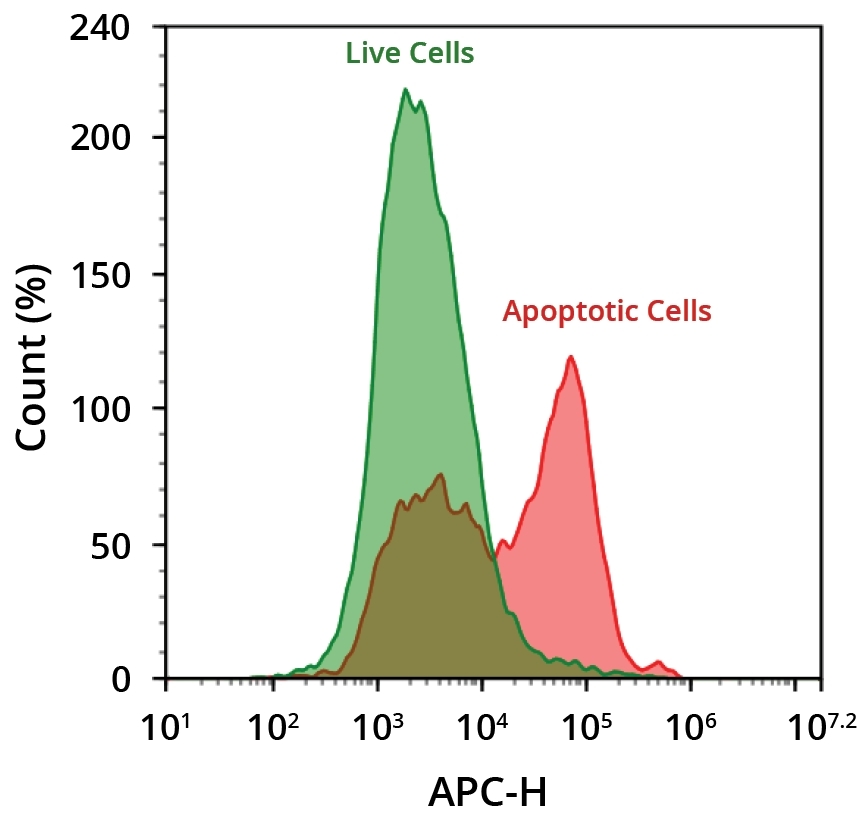
AAT Bioquest's iFluor® dyes are optimized for labeling proteins, in particular, antibodies. These dyes are optimized to have minimal fluorescence quenching effect on proteins and nucleic acids. Our iFluor® 680 dyes have fluorescence excitation and emission maxima close to 680 nm and 700 nm respectively with good photostability. These spectral characteristics make them an excellent alternative to Cy5.5®, IRDye® 700 and Alexa Fluor® 680 (Cy5.5®, IRDye® and Alexa Fluor® are the trademarks of GE Healthcare, Li-COR and Invitrogen respectively). ReadiLink™ labeling kits essentially only require 2 simple mixing steps without a column purification needed. iFluor® 680 SE used in this ReadiLink™ kit is reasonably stable and shows good reactivity and selectivity with protein amino groups. The kit has all the essential components for labeling ~2x50 ug antibody. Each of the two vials of iFluor® 680 dye provided in the kit is optimized for labeling ~50 µg antibody. iFluor® 680 SE protein labeling kit provides a convenient method to label monoclonal, polyclonal antibodies or other proteins (>10 kDa) with the iFluor® 680 SE.
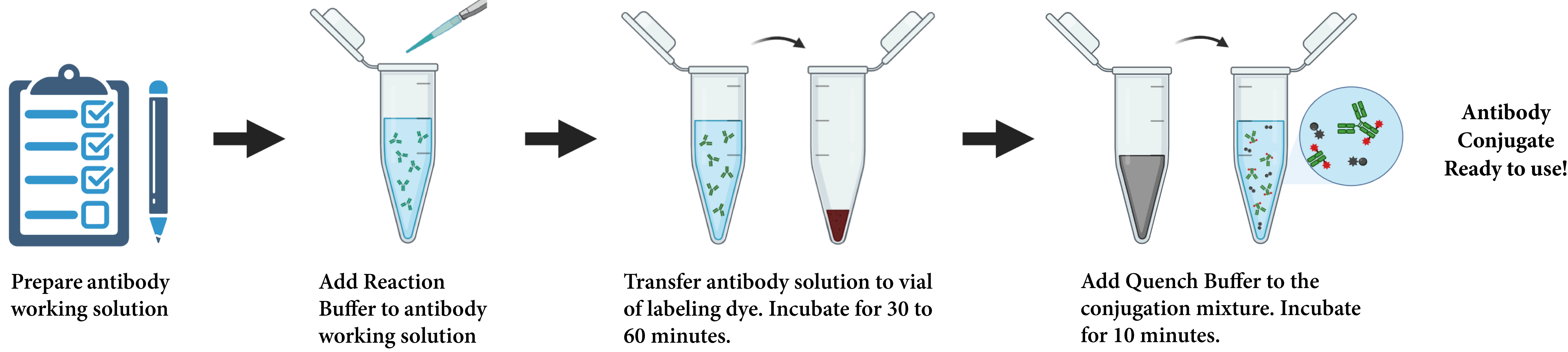
Figure 1. Overview of the ReadiLink™ Rapid Antibody Labeling protocol. In just two simple steps, and with no purification necessary, covalently label microgram amounts of antibodies in under an hour.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1240 | 2 Labelings | Price |
Spectral properties
| Correction factor (260 nm) | 0.097 |
| Correction factor (280 nm) | 0.094 |
| Extinction coefficient (cm -1 M -1) | 220000 1 |
| Excitation (nm) | 684 |
| Emission (nm) | 701 |
| Quantum yield | 0.23 1 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 20, 2026

