ReadiUse™ PE
R-Phycoerythrin; Ammonium Sulfate-Free
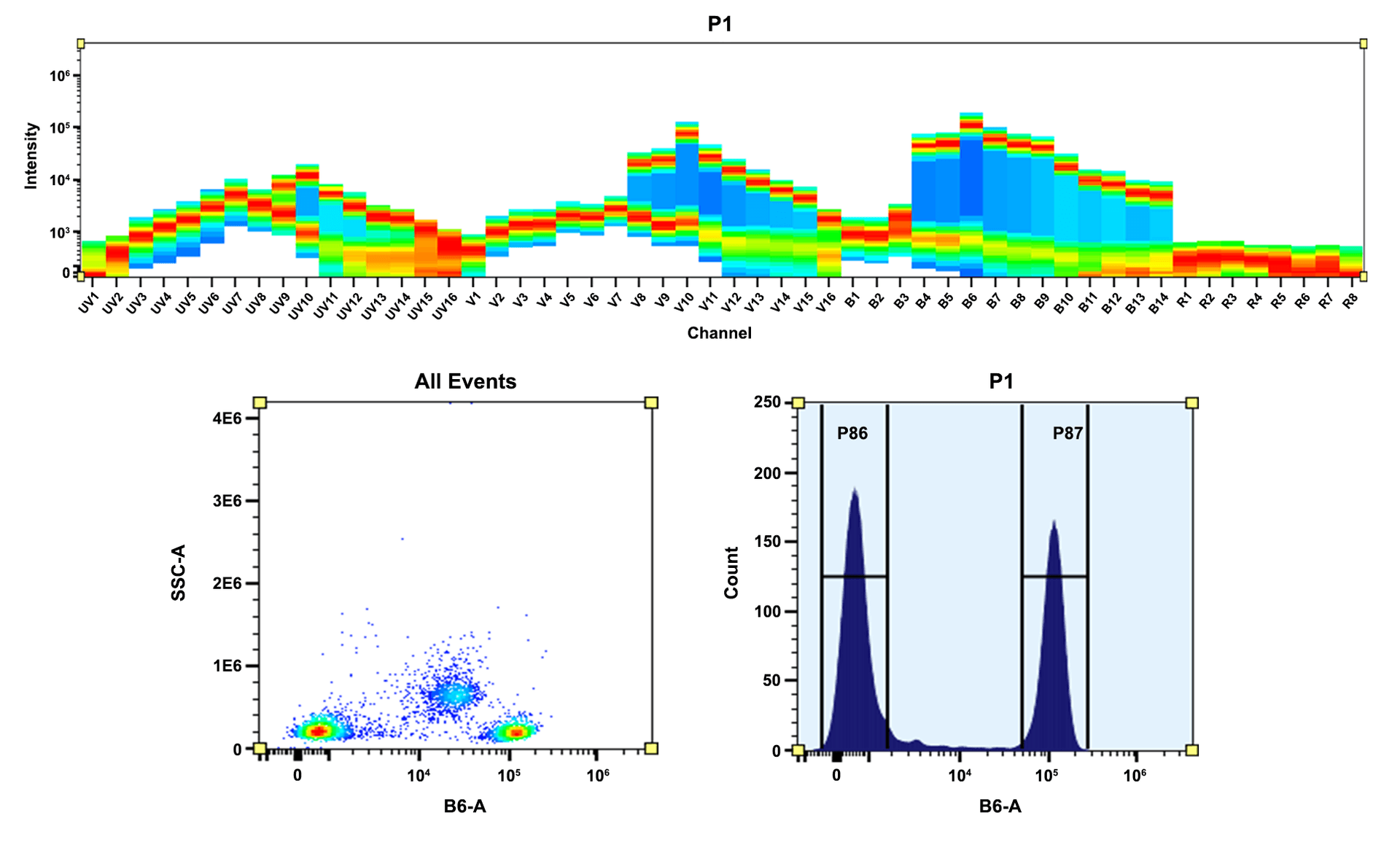
R-Phycoerythrin (PE) is isolated from red algae. Its primary absorption peak is at 565 nm with secondary peaks at 496 and 545 nm. All the commercial PE materials are sold in concentrated ammonium sulfate buffers. The commercial PE materials from other vendors require the tedious dialysis or other purifications performed before it can be used for labeling purposes. AAT Bioquest offers this ReadiUse™ PE that can be readily used for any labelings without any purifications required. Our highly purified ReadiUse™ PE facilitates the rapid PE conjugations to antibodies and other proteins such as streptavidin and other secondary reagents.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 2500 | 1 mg | Price | |
| 2501 | 10 mg | Price |
Physical properties
| Molecular weight | ~240000 |
| Solvent | Water (Add 100 µL of H₂O to Cat# 2500 or 1 mL to Cat# 2501 to reconstitute to 10 mg/mL in PBS) |
Spectral properties
| Correction factor (280 nm) | 0.175 |
| Extinction coefficient (cm -1 M -1) | 1960000 |
| Excitation (nm) | 565 |
| Emission (nm) | 574 |
| Quantum yield | 0.82 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Refrigerated (2-8 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 23, 2025

