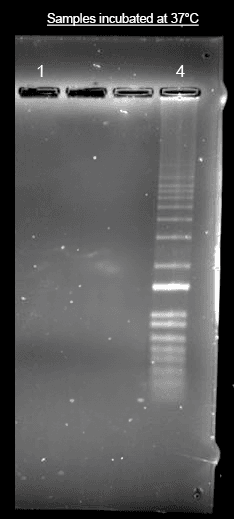
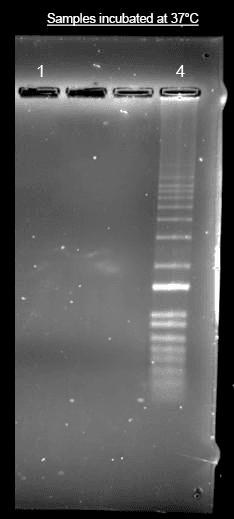
Helixyte™ iFluor® 350 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA*
| Price | |
| Catalog Number | |
| Unit Size | |
| Quantity |
| Telephone | 1-800-990-8053 |
| Fax | 1-800-609-2943 |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Shipping | Standard overnight for United States, inquire for international |
| Solvent | DMSO |
| Correction Factor (260 nm) | 0.83 |
| Correction Factor (280 nm) | 0.23 |
| Extinction coefficient (cm -1 M -1) | 200001 |
| Excitation (nm) | 345 |
| Emission (nm) | 450 |
| Quantum yield | 0.951 |
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| Overview |
Correction Factor (260 nm) 0.83 | Correction Factor (280 nm) 0.23 | Extinction coefficient (cm -1 M -1) 200001 | Excitation (nm) 345 | Emission (nm) 450 | Quantum yield 0.951 |
Example protocol
AT A GLANCE
Combine DNA with the Helixyte™ iFluor® 350 Nucleic Acid Labeling Dye stock solution.
Incubate for 1 hour at 37°C.
Purify the conjugate as required for downstream applications.
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Before opening the vial, thaw Helixyte™ iFluor® nucleic acid labeling dye at room temperature. Briefly centrifuge to collect the dried pellet.
Add 110 μL of DMSO to the Helixyte™ iFluor® 350 Nucleic Acid Labeling Dye vial to prepare a 10 mM stock solution.
Note: It is recommended to divide any unused stock solution into single-use aliquots. Store the aliquots at ≤-20 ºC and protect them from light. Avoid repeated freeze-thaw cycles.
SAMPLE EXPERIMENTAL PROTOCOL
Prepare the labeling reaction according to the specifications in table 1 below.
Table 1. Standard Nucleic Acid Labeling Reaction.
Components Volume added to reaction Final Concentration DNA (1 mg/mL) 2 to 5 µL 2 to 5 µg Helixyte™ iFluor® 350 Nucleic acid Labeling Dye stock solution 0.5 µL 50 µM TE Buffer (pH 8 to 8.5) Add sufficient buffer to adjust the volume to 100 µL Note: This DNA:Dye ratio results in labeling efficiencies that are appropriate for most applications. The amount of Helixyte™ iFluor® 350 Nucleic Acid Labeling Dye or the reaction incubation time can be adjusted to modify the labeling density as per the application requirements. The DNA-to-dye ratio must be optimized to achieve a higher labeling ratio
Incubate the reaction at 37℃ for 1 hour, protected from light.
Note: After 30 minutes of incubation, briefly centrifuge the reaction to minimize the effects of evaporation and maintain the appropriate concentration of the reaction components.
Note: Alternatively, the reaction can be incubated at room temperature for 2 hours. For the best labeling condition, we recommend incubating at 37℃.
After incubation, the labeling mix can be purified to remove any free labeling dye. Refer to the “Purification of labeling mix with alcohol precipitation” section below for instructions.
Add 0.1 volume (10 uL) of 5M sodium chloride and 2 - 2.5 volumes of ice-cold 100% ethanol (250 uL) to the reaction. Mix well and place at ≤ -20°C for at least 30 minutes.
Centrifuge at full speed (>14,000 x g) in a refrigerated micro centrifuge for 15-30 minutes to pellet the labeled nucleic acid. Once pelleted, carefully remove the ethanol with a micropipette. Do not disturb the pellet.
Note: Small nucleic acid quantities can be difficult to visualize. Mark and orient the precipitate-containing tubes in the microfuge such that the pellet will form in a predetermined place.
Wash the pellet once with 500 μL of room temperature 70% ethanol. Centrifuge at full speed for an additional 15-30 minutes.
Remove all traces of ethanol with a micropipette. DO NOT allow the sample to dry longer than 5 minutes as the pellet may become difficult to resuspend.
Resuspend the labeled DNA with ~ 30 µL sterile water.
Store the purified, labeled nucleic acid for long-term storage or put on ice for immediate use.
Spectrum
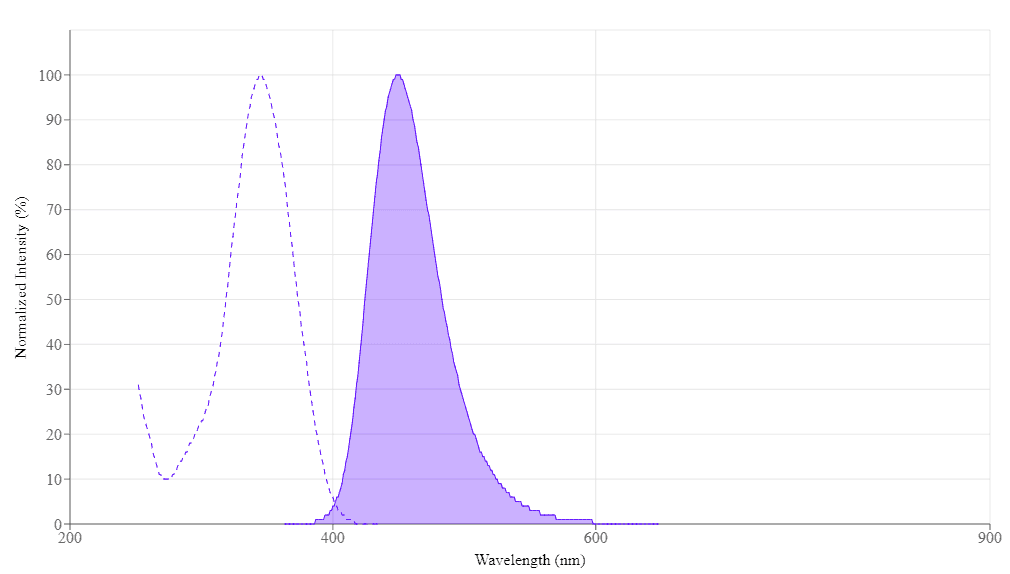
Spectral properties
| Correction Factor (260 nm) | 0.83 |
| Correction Factor (280 nm) | 0.23 |
| Extinction coefficient (cm -1 M -1) | 200001 |
| Excitation (nm) | 345 |
| Emission (nm) | 450 |
| Quantum yield | 0.951 |
Product Family
| Name | Excitation (nm) | Emission (nm) | Extinction coefficient (cm -1 M -1) | Quantum yield | Correction Factor (260 nm) | Correction Factor (280 nm) |
| Helixyte™ iFluor® 488 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 491 | 516 | 750001 | 0.91 | 0.21 | 0.11 |
| Helixyte™ iFluor® 555 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 557 | 570 | 1000001 | 0.641 | 0.23 | 0.14 |
| Helixyte™ iFluor® 594 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 587 | 603 | 2000001 | 0.531 | 0.05 | 0.04 |
| Helixyte™ iFluor® 647 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 656 | 670 | 2500001 | 0.251 | 0.03 | 0.03 |
| Helixyte™ iFluor® 750 Nucleic Acid Labeling Dye *Optimized for Labeling 2x100 ug DNA/RNA* | 757 | 779 | 2750001 | 0.121 | 0.044 | 0.039 |
Images
References
Authors: Madireddy, Advaitha and Gerhardt, Jeannine
Journal: STAR protocols (2023): 102721
Authors: Wang, Yong and Wang, Dandan and Qi, Guohua and Hu, Ping and Wang, Erkang and Jin, Yongdong
Journal: Analytical chemistry (2023): 16234-16242
Authors: Satusky, Matthew J and Johnson, Caitlin V and Erie, Dorothy A
Journal: Biophysical journal (2023): 1211-1218
Authors: Mittal, Sneha and Pathak, Biswarup
Journal: ACS applied bio materials (2023): 218-227
Authors: Ma, Ningning and Liu, Haijing and Wu, Yaqian and Yao, Mengfei and Zhang, Bo
Journal: International journal of molecular sciences (2022)
Authors: Lyu, Ruitu and Wu, Tong and Zhu, Allen C and West-Szymanski, Diana C and Weng, Xiaocheng and Chen, Mengjie and He, Chuan
Journal: Nature protocols (2022): 402-420
Authors: Ostersehlt, Lynn M and Jans, Daniel C and Wittek, Anna and Keller-Findeisen, Jan and Inamdar, Kaushik and Sahl, Steffen J and Hell, Stefan W and Jakobs, Stefan
Journal: Nature methods (2022): 1072-1075
Authors: Sampieri, Alicia and Monroy-Contreras, Ricardo and Asanov, Alexander and Vaca, Luis
Journal: Frontiers in bioengineering and biotechnology (2022): 881679
Authors: Bai, Min and Cao, Xiaowen and Chen, Feng and Xue, Jing and Zhao, Yue and Zhao, Yongxi
Journal: Analytical chemistry (2021): 10495-10501
Authors: Chen, Kaikai and Gularek, Felix and Liu, Boyao and Weinhold, Elmar and Keyser, Ulrich F
Journal: ACS nano (2021): 2679-2685