A Superior Direct Method for the Quantitative Analysis of DNA Fragments in Late-Stage Apoptosis
Multicellular organisms are made up of highly organized community of cells. For this community to thrive, it is important to establish homeostasis through complex cellular processes such as cell proliferation and apoptosis. The inappropriate shift in favor of either process has been implicated to cause disease. For instance, an increase in apoptotic activity in neurons is the source of various neurodegenerative disorders such as Alzheimer's and Parkinson's disease. In contrast, the loss of apoptotic mechanisms can lead to excessive cellular growth which is an underlying mechanism of how cells become cancerous. Overall, the role of apoptosis in disease is of high interest in scientific research.
Apoptosis
Apoptosis is a pathway of sophisticated and well-orchestrated programmed cell death. This cell-autonomous process is characterized by several distinct morphological and biochemical markers including cell and nuclear shrinkage, chromatin condensation, cytoplasmic membrane blebbing, and nuclear DNA fragmentation. These morphological changes all stem from two apoptotic pathways: the extrinsic or the intrinsic pathway. The extrinsic pathway is a receptor-mediated pathway activated by extracellular death receptor ligands (e.g. tumor necrosis factor (TNF) or Fas ligand (FasL)). The intrinsic pathway is induced intracellularly by environmental stressors resulting in mitochondrial membrane permeabilization and the release of apoptosis inducing factor (AIF) and cytochrome c. Signals from both pathways are propagated through the caspase cascade, which ultimately lead to the downstream digestion of nuclear DNA by caspase activated nucleases. These features of apoptosis allow for the ability to use DNA fragmentation assays as the ultimate determinate for identifying apoptosis.
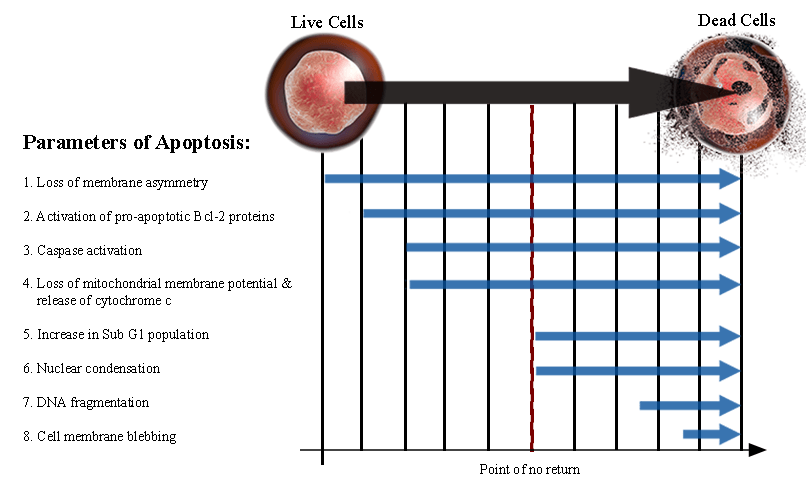
Outlines all of the parameters involved in apoptosis. It is important to note that these events do not occur in order. Many of them will overlap and will often occur simultaneously. Events 1 through 4 are biochemical features of apoptosis which do not necessarily end in cell death. Downstream apoptotic events 5 through 8 are strong indicators that a cell has committed to apoptosis, after which it is very unlikely the cell will survive.
Apoptotic assays detecting DNA fragmentation
There are various techniques for the assessment and identification of apoptotic DNA fragmentation. The earliest assay is a gel-based DNA laddering assay which involved using agarose gel electrophoresis to visualize and separate DNA fragments. Apoptotic DNA fragmentation is indicated by a distinctive 'ladder' like pattern of nucleosomal fragments approximately 180-200 base pairs in length. Although effective at generating qualitative data, this technique is not easily adaptable for high throughput use.
Another method of detection employs a double-antibody sandwich ELSIA assay consisting of anti-DNA and anti-histone antibodies to visualize DNA fragmentation during apoptosis. Although this assay is significantly more sensitive at detecting apoptotic DNA fragments and amenable for high-throughput uses (e.g. compound screening), it is labor intensive and the use of antibodies is costly.
TUNEL assay
In 1992, Gorczyca et al. 1992 introduced a new method of measuring apoptotic DNA fragmentation utilizing terminal deoxynucleotidyl transferase (TdT) and nick translation or TUNEL assay. TdT catalyzes the addition of a fluorophore-labeled deoxyuridine triphosphate (dUTP) to the 3'-hydroxyl termni of a DNA double strand break (DSB).
Since the introduction of the TUNEL assay, it has become a widely-accepted in situ technique for identifying apoptotic cell death. Compared to previous methods, the TUNEL assay is more sensitive with the capacity to give a broad range of quantitative measurements over several orders of magnitude. Over the years, the TUNEL technique has been adapted and optimized for chromogenic and fluorogenic detection methods. Chromogenic TUNEL assays utilize dUTP conjugated to biotin and an enzyme-labeled streptavidin for high sensitive staining. The fluorogenic detection method relies on fluorescent dye-conjugated dUTP which is used for direct detection of DNA fragmentation.
Several variables influence the staining kinetics of TUNEL assays. Some of these variables include reagent concentration, fixation of sample, and accessibility of DNA strand breaks, which may vary between tissue or cell sample types. Therefore, standardizing your chosen TUNEL assay and having appropriate controls are critical in data collection and interpretation.
Cell Meter™ TUNEL apoptosis assay
AAT Bioquest's Cell Meter™ TUNEL Apoptosis Assay is optimized for fluorogenic detection and is available in either green or red fluorescence emission colors. AAT Bioquest's Cell Meter™ Assays include all the essential components and a robust protocol to detect and quantitate apoptotic DNA fragmentation in cells. Measuring apoptotic DNA fragments is achieved by catalytically incorporating a fluorescent dye modified deoxyuridine 5'-triphosphate nucleotide, Tunnelyte™ Green or Tunnelyte™ Red, to the 3' OH ends of DNA using TdT. Labeled DNA fragments can then be quantitated by flow cytometry or directly visualized by fluorescence microscopy.
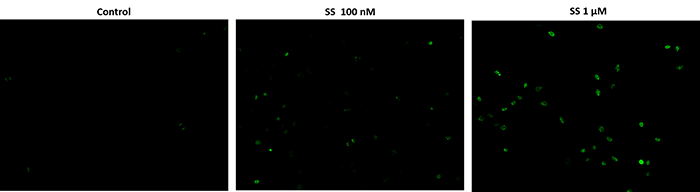
Fluorescence images of TUNEL reaction in HeLa cells treated with a chemical that induces apoptosis, staurosporin (SS). The HeLa cells are treated with 100 nM or 1 µM SS for 4 hours as compared to the untreated control. Cells were incubated with reaction mixture for 1 hour at 37°C. The green fluorescence signal was analyzed using fluorescence microscope with a FITC filter set. Fluorescently labeled DNA strand breaks shows intense fluorescent staining in SS treated cells.
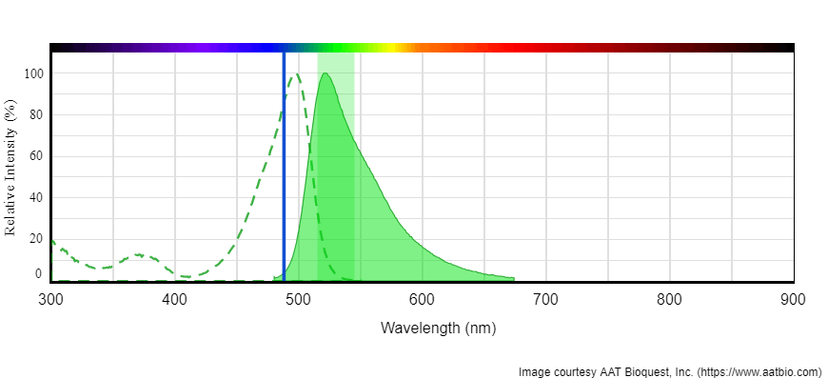
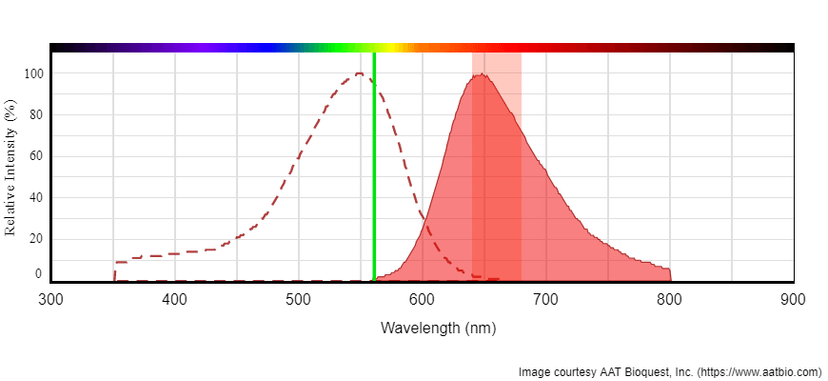
Tunnelyte™ Green's peak absorption at 497 nm is well-suited for efficient excitation by instruments equipped with the 488 nm line of an argon-ion laser. Tunnelyte™ Red's peak absorption at 547 nm is efficiently excited with instruments using the helium-neon laser. For emission filter selection, Tunnelyte™ Green or Tunnelyte™ Red can be read using a FITC (530/30) or Cy5 (660/40) filter, respectively. Because Tunnelyte™ nucleotides are amply separated from the blue spectrum, stains such as DAPI, Hoechest or Nuclear Violet™ LCS1are suitable for live cell counterstaining in multicolor fluorescence applications.
Left: Absorption and emission spectrum for Tunnelyte™ Green. Excitation is at 488 nm (Blue laser line). Tunnelyte™ Green read with emission filter FITC (Green band). Right: Absorption and emission spectrum of Tunnelyte™ Red. Excitation is at 561 nm (Green laser line). Tunnelyte™ Red read with emission filter Cy5 (Red band).
Our Cell Meter™ TUNEL Apoptosis assay offers a distinct advantage over other commercially available TUNEL assays, because our kits do not contain sodium or potassium cacodylate in the reaction buffer. Elimination of sodium cacodylate has a two-fold benefit. First, it is much safer to handle. Second, it provides a more sensitive and accurate apoptosis assay. Sodium cacodylate is a carcinogenic arsenic derivative that is highly toxic by ingestion, inhalation or skin contact. Because of the toxicity, it can induce apoptosis which inevitably leads to noise when measuring apoptosis in the assay.
The average market price and unit size of commercially available TUNEL assays is $529 for 45 tests. AAT Bioquest's Cell Meter™ TUNEL assays offer the best value with each kit priced at $327 and a unit size of 50 tests.
Table 1. Comparison of TUNEL assays
| Company ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ | Price ▲ ▼ |
| AAT Bioquest | Cell Meter™ Live Cell TUNEL Apoptosis Assay Kit *Green Fluorescence* | 50 Tests | $334 |
| ThermoFisher | Click-iT™ TUNEL Alexa Fluor™ 488 Imaging Assay | 1 Kit (50 coverslips) | $860 |
| Promega | DeadEnd™ Fluorometric TUNEL System | 60 Reactions | $597 |
| Abcam | TUNEL Assay Kit " In situ BrdU-RED DNA Fragmentation | 60 Tests | $735 |
Conclusion
AAT Bioquest's Cell Meter™ TUNEL assay kits offers a simple, safe, and sensitive approach for detecting apoptotic DNA fragmentation. The direct incorporation of fluorescent nucleotides cuts down on assay time by reducing the number required incubation steps which allows for a quicker labeling of fragmented DNA. Finally our reagents are free of sodium cacodylate which results in the easier handling and disposal of the waste generated with our TUNEL assay.
Table 2. Cell Meter Tunel Assays
| Cat No. ▲ ▼ | Product Name ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ |
| 22849 | Cell Meter™ Live Cell TUNEL Apoptosis Assay Kit *Green Fluorescence* | 498 | 522 |
| 22844 | Cell Meter™ Live Cell TUNEL Apoptosis Assay Kit *Red Fluorescence* | 549 | 648 |
References
- Darzynkiewicz, Z., et al. "Features of Apoptotic Cells Measured by Flow Cytometry."Cytometry, vol. 13, no. 8, 1992, pp. 795"808., doi:10.1002/cyto.990130802.
- Darzynkiewicz, Zbigniew, et al. "Analysis of apoptosis by cytometry using TUNEL assay." Methods, vol. 44, no. 3, 2008, pp. 250"254., doi:10.1016/j.ymeth.2007.11.008.
- Kerr, J F R, et al. "Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics." British Journal of Cancer, vol. 26, no. 4, 1972, pp. 239"257., doi:10.1038/bjc.1972.33.
- Liu, Xuesong, et al. "DFF, a Heterodimeric Protein That Functions Downstream of Caspase-3 to Trigger DNA Fragmentation during Apoptosis." Cell, vol. 89, no. 2, 1997, pp. 175"184., doi:10.1016/s0092-8674(00)80197-x.
- Lu, Z., et al. "Nucleoplasmin regulates chromatin condensation during apoptosis." Proceedings of the National Academy of Sciences, vol. 102, no. 8, July 2005, pp. 2778"2783., doi:10.1073/pnas.0405374102.
- Tang, Damu, and Vincent J. Kidd. "Cleavage of DFF-45/ICAD by Multiple Caspases Is Essential for Its Function during Apoptosis." Journal of Biological Chemistry, vol. 273, no. 44, 1998, pp. 28549"28552., doi:10.1074/jbc.273.44.28549.