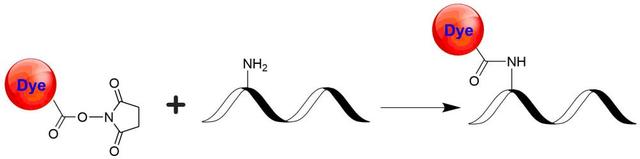
trFluor™ Eu succinimidyl ester *europium complex*
Example protocol
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Mix 100 µL of a reaction buffer (e.g., 1 M sodium carbonate solution or 1 M phosphate buffer with pH ~9.0) with 900 µL of the target protein solution (e.g., antibody, protein concentration >2 mg/mL if possible) to give 1 mL protein labeling stock solution.
Note: The pH of the protein solution (Solution A) should be 8.5 ± 0.5. If the pH of the protein solution is lower than 8.0, adjust the pH to the range of 8.0-9.0 using 1 M sodium bicarbonate solution or 1 M pH 9.0 phosphate buffer.
Note: The protein should be dissolved in 1X phosphate-buffered saline (PBS), pH 7.2-7.4. If the protein is dissolved in Tris or glycine buffer, it must be dialyzed against 1X PBS, pH 7.2-7.4, to remove free amines or ammonium salts (such as ammonium sulfate and ammonium acetate) that are widely used for protein precipitation.
Note: Impure antibodies or antibodies stabilized with bovine serum albumin (BSA) or gelatin will not be labeled well. The presence of sodium azide or thimerosal might also interfere with the conjugation reaction. Sodium azide or thimerosal can be removed by dialysis or spin column for optimal labeling results.
Note: The conjugation efficiency is significantly reduced if the protein concentration is less than 2 mg/mL. For optimal labeling efficiency, the final protein concentration range of 2-10 mg/mL is recommended.
Add anhydrous DMSO into the vial of trFluor™ Eu succinimidyl ester to make a 10 mM stock solution. Mix well by pipetting or vortex.
Note: Prepare the dye stock solution (Solution B) before starting the conjugation. Use promptly. Extended storage of the dye stock solution may reduce the dye activity. Solution B can be stored in the freezer for two weeks when kept from light and moisture. Avoid freeze-thaw cycles.
SAMPLE EXPERIMENTAL PROTOCOL
This labeling protocol is specifically optimized for the conjugation of goat anti-mouse IgG with trFluor™ Eu succinimidyl ester. Further optimization maybe required for other proteins.
Note: Each protein requires a distinct dye/protein ratio, which also depends on the properties of dyes. Over-labeling of a protein could detrimentally affect its binding affinity while the protein conjugates of low dye/protein ratio give reduced sensitivity.
Use a 10:1 molar ratio of Solution B (dye)/Solution A (protein) as a starting point. Add 5 µL of the dye stock solution (Solution B, assuming the dye stock solution is 10 mM) into the vial of the protein solution (95 µL of Solution A) with effective shaking. The concentration of the protein is ~0.05 mM assuming the protein concentration is 10 mg/mL and the molecular weight of the protein is ~200KD.
Note: We recommend using a 10:1 molar ratio of Solution B (dye)/Solution A (protein). If it is too low or too high, determine the optimal dye/protein ratio at 5:1, 15:1, and 20:1, respectively.
- Continue to rotate or shake the reaction mixture at room temperature for 30-60 minutes.
The following protocol is an example of dye-protein conjugate purification by using a Sephadex G-25 column.
Prepare Sephadex G-25 column according to the manufacture instruction.
Load the reaction mixture (From "Run conjugation reaction") to the top of the Sephadex G-25 column.
Add PBS (pH 7.2-7.4) as soon as the sample runs just below the top resin surface.
Add more PBS (pH 7.2-7.4) to the desired sample to complete the column purification. Combine the fractions that contain the desired dye-protein conjugate.
Note: For immediate use, the dye-protein conjugate needs to be diluted with staining buffer, and aliquoted for multiple uses.
Note: For longer-term storage, the dye-protein conjugate solution needs to be concentrated or freeze-dried.
Calculators
Common stock solution preparation
| 0.1 mg | 0.5 mg | 1 mg | 5 mg | 10 mg | |
| 1 mM | 58.628 µL | 293.142 µL | 586.283 µL | 2.931 mL | 5.863 mL |
| 5 mM | 11.726 µL | 58.628 µL | 117.257 µL | 586.283 µL | 1.173 mL |
| 10 mM | 5.863 µL | 29.314 µL | 58.628 µL | 293.142 µL | 586.283 µL |
Molarity calculator
| Mass (Calculate) | Molecular weight | Volume (Calculate) | Concentration (Calculate) | Moles | ||||
| / | = | x | = |
Spectrum
Product family
| Name | Excitation (nm) | Emission (nm) | Correction Factor (260 nm) | Correction Factor (280 nm) |
| trFluor™ Tb succinimidyl ester | 333 | 544 | 0.942 | 0.797 |
References
Authors: Lo MC, Ngo R, Dai K, Li C, Liang L, Lee J, Emkey R, Eksterowicz J, Ventura M, Young SW, Xiao SH.
Journal: Anal Biochem (2012): 368
Authors: Saville L, Spais C, Mason JL, Albom MS, Murthy S, Meyer SL, Ator MA, Angeles TS, Husten J.
Journal: Assay Drug Dev Technol. (2012)
Authors: Paila YD, Kombrabail M, Krishnamoorthy G, Chattopadhyay A.
Journal: J Phys Chem B (2011): 11439
Authors: Martikkala E, Rozw and owicz-Jansen A, Hanninen P, Petaja-Repo U, Harma H.
Journal: J Biomol Screen (2011): 356
Authors: Gaborit N, Larbouret C, Vallaghe J, Peyrusson F, Bascoul-Mollevi C, Crapez E, Azria D, Chardes T, Poul MA, Mathis G, Bazin H, Pelegrin A.
Journal: J Biol Chem (2011): 11337