Amplite® Fluorimetric NADP Assay Kit
Blue Fluorescence
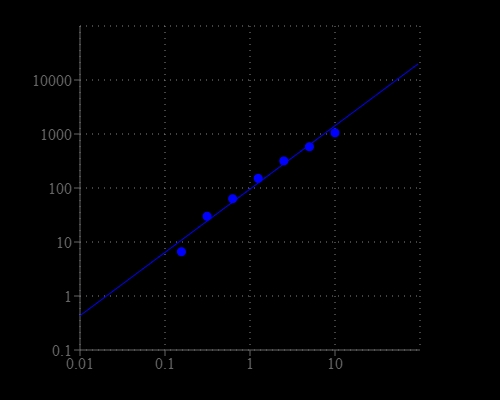
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) are two important cofactors for many enzyme reactions found in living cells. NAD forms NADP with the addition of a phosphate group to the 2' position of the adenyl nucleotide through an ester linkage. NADP is used in anabolic biological reactions, such as fatty acid and nucleic acid synthesis, which requires NADPH as a reducing agent. In chloroplasts, NADP is an oxidizing agent important in the preliminary reactions of photosynthesis. The NADPH produced by photosynthesis is used as reducing power for the biosynthetic reactions in the Calvin cycle of photosynthesis. Quantifying the generation or consumption of these factors is an important method to monitor the enzyme-mediated reaction or screening the modulator or substrate of these enzyme reactions. There are several kits on the market to quantify NADPH or total NADP/NADPH amount, but detection NADP generation in the presence of large excess amount of NADPH has been quite challenging to date because NADP has its absorption peak at 260 nm and does not fluorescence, making the measurement unpractical. Amplite® Fluorimetric NADP Assay Kit provides a sensitive and rapid detection of NADP. The kit directly measure NADP using Quest Fluor™ NADP reagent, our newly developed NADP sensor. The proprietary probe used in this kit reacts only with NADP to generate a product that fluorescence at a specific excitation and emission spectra range and has little response to NADPH. This kit can detect as little as 30 nM NADP in a 100 µL assay volume, and monitor 0.3% NADP generation in the presence of excess amount of NADPH. This assay can be performed in a convenient 96-well or 384-well microtiter-plate format and can be used in high-throughput screening.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 15281 | 200 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 420 nm |
| Emission | 480 nm |
| Cutoff | 455 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 24, 2026
