Cell Meter™ Cellular Senescence Activity Assay Kit
Green Fluorescence
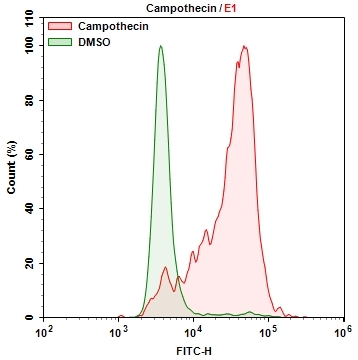
Cellular Senescence is an irreversible growth arrest triggered in order to prevent growth in DNA damaged cells. Senescence-associated beta-galactosidase (SA-beta-gal) is highly overexpressed in senescent cells and it has been widely used as a senescence marker. X-gal staining, a colorimetric method is widely available and used to detect SA-beta-gal in senescent cells. The color method has some limitations such as requirement of fixation of samples due to the low cell permeability of X-gal, longer staining time and low sensitivity. Cell Meter™ Cellular Senescence Activity Assay Kit uses Xite™ beta-D-galactopyranoside, a fluorogenic beta-Gal substrate that readily enters into live cells, and gets cleaved by SA-β-gal inside cells, generating strong green fluorescence. Unlike cell-impermeable X-Gal substrate, it has excellent cell permeability. Cell Meter™ Cellular Senescence Activity Assay Kit enables users to detect the senescence with higher sensitivity with robust performance. The Xite product is well retained inside the cells, producing a stable signal for fluorescence imaging and flow cytometry analysis.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 23005 | 100 Tests | Price |
Spectral properties
| Absorbance (nm) | 487 |
| Correction factor (260 nm) | 0.32 |
| Correction factor (280 nm) | 0.35 |
| Extinction coefficient (cm -1 M -1) | 80000 1 |
| Excitation (nm) | 498 |
| Emission (nm) | 517 |
| Quantum yield | 0.7900 1 , 0.952 |
Storage, safety and handling
| Intended use | Research Use Only (RUO) |
Instrument settings
| Flow cytometer | |
| Excitation | 488 nm laser |
| Emission | 530/30 nm filter |
| Instrument specification(s) | FITC channel |
| Fluorescence microscope | |
| Excitation | FITC filter set |
| Emission | FITC filter set |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 26, 2026

