CytoTrace™ Ultra Green
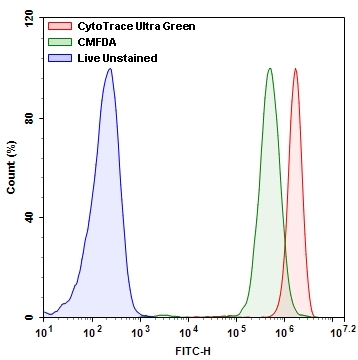
CMFDA (5-chloromethylfluorescein diacetate) has been used as a prominent cell tracker in the research community for a long time. We are introducing a new generation of fluorescent dye, named CytoTrace™ Ultra Green that is well suited for tracking the cell movement or location. The dye is well retained in cells, allowing tracking live cells for quite a few generations. Compared to CMFDA under the same conditions, CytoTrace™ Ultra Green is significantly brighter, more photostable and robust to use. Its fluorescence signal is also fixable. The signal generated by this dye is highly stable after fixation, making it ideal candidate to combine with other types of assays such as cytotoxicity etc for multicolor applications. The excitation and emission spectra of CytoTrace™ Ultra Green is identical to those of FITC, and are well separated from the common red fluorescent dyes such as Texas Red, Cy5, Cy7, iFluor® 647 & 750, Alexa Fluor® 647 and 750. CytoTrace™ Ultra Green can be readily used for tracking live cells for various biological applications, and compatible with flow cytometry and fluorescence microscopy.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 21800 | 1 mg | Price |
Physical properties
| Molecular weight | 822.72 |
| Solvent | DMSO |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12352200 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 7, 2026
