Helixyte™ Green Fluorimetric Total Nucleic Acid Quantitation Kit
Optimized for Microplate Readers
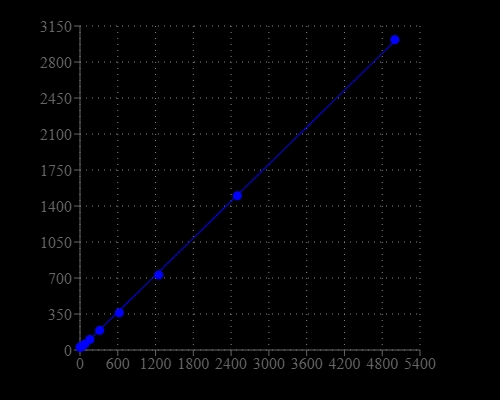
Helixyte™ Green Fluorimetric Total Nucleic Acid Quantitation Kit is designed to measure total amounts of nucleic acids, including double-stranded DNA (dsDNA), single-stranded DNA (ssDNA) and RNA in an easy and accurate format. The kit has all the essential reagents, including Helixyte™ Green All reagent, dilution buffer, and pre-diluted DNA standards. Helixyte™ Green All reagent is a sensitive fluorescent nucleic acid probe for measuring the total amounts of nucleic acids in a sample that may contain dsDNA, ssDNA, RNA and long oligonucleotides. Helixyte™ Green All reagent indiscriminately binds to dsDNA, ssDNA and RNA. Helixyte™ Green Fluorimetric Total Nucleic Acid Quantitation Kit is optimized for measuring the total amounts of nucleic acids with a fluorescence microplate reader.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17630 | 200 Tests | Price |
Spectral properties
| Excitation (nm) | 509 |
| Emission (nm) | 527 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12171501 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 24, 2026

