Hoechst 33342
Ultrapure Grade
Hoechst 33342 Ultrapure Grade is a cell-permeable blue fluorescent dye that binds to AT-rich regions in the minor groove of DNA, enabling live cell nuclear staining, cell cycle analysis, and identification of side population stem cells.
- Optimal UV Brightness: Maximum fluorescence under UV or near-UV excitation for clear nuclear visualization
- Low Background Noise: Minimal cytoplasmic staining ensures clean nuclear segmentation in imaging applications
- Live-Cell Compatible: Cell-permeable design allows real-time nuclear morphology and cell cycle studies
- Sigma/Invitrogen Equivalent: Industry-standard performance with enhanced purity and batch-to-batch consistency

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17530 | 100 mg | Price | |
| 17533 | 1 g | Price |
Overview
Hoechst 33342 is a cell membrane-permeant, fluorescent DNA stain acclaimed for its strong binding affinity to A–T-rich regions in the minor groove of double-stranded DNA. This dye surpasses many nuclear stains due to its additional ethyl substituent, which boosts membrane permeability—thus seamlessly crossing the plasma membrane of live cells. As a result, Hoechst 33342 is widely utilized in diverse fields such as cell biology, immunology, neuroscience, and high-throughput drug discovery.
At AAT Bioquest, we deliver easy-to-use Hoechst 33342 solutions (equivalent to formulations from Sigma or Invitrogen). Built on decades of fluorescence chemistry expertise, our brand guarantees:
- Optimal brightness under UV or near-UV excitation
- Low background noise for clean nuclear segmentation
- High consistency across batches due to rigorous QC and thorough purification
Essential Properties
Chemical Family & Basic Properties
- Chemical Family: Bisbenzimide (minor groove-binding)
- Molecular Weight: 561.93 g/mol
Note on Membrane Permeability: Structurally, Hoechst 33342 has an extra ethyl group for high membrane permeability (Kirby et al., 2024). Once bound to DNA, it emits intense blue fluorescence—ideal for a wide range of imaging and flow cytometry applications.
Excitation & Emission Characteristics
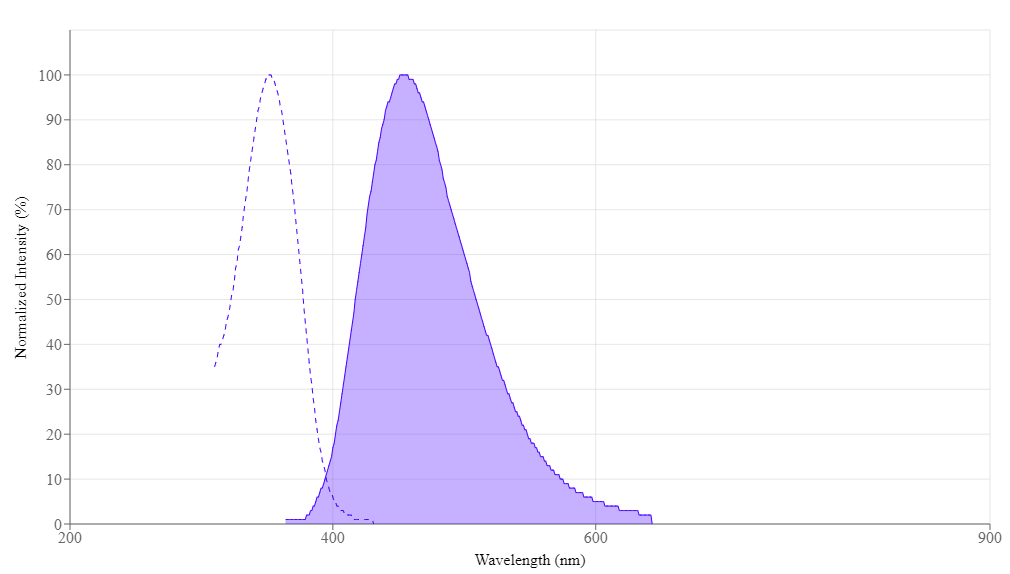
- Excitation Maximum: ~350–355 nm (UV or near-UV lasers)
- Emission Maximum: ~461 nm (bright blue region)
Its spectral overlap with DAPI allows reuse of existing filter sets. Flow cytometers with a 355 nm or 405 nm laser readily detect Hoechst 33342.
Working Concentration & Dosing Calculations
Given its 561.93 g/mol molecular weight, a 20 mM stock translates to ~11.2 mg/mL. Accurate calculations ensure reproducible staining intensities across experiments.
Given its 561.93 g/mol molecular weight, a 20 mM stock translates to ~11.2 mg/mL. Accurate calculations ensure reproducible staining intensities across experiments.
Key Advantages
- Enhanced Membrane Permeability vs. Hoechst 33258 and DAPI
- Bright, Stable Fluorescence in the blue emission channel (~461 nm)
- High Compatibility with standard immunofluorescence panels
- Reduced Photobleaching relative to older UV-excited dyes
- Versatility in Live or Fixed Cells for both real-time and endpoint assays
Hoechst 33342 vs. DAPI
- Hoechst 33342: More lipophilic, suitable for real-time studies, relatively low toxicity
- DAPI: More toxic for live cells, typically used for fixed tissues or endpoint analysis
Mechanism of Action: Minor Groove Binding
Hoechst 33342 is a non-intercalating dye that binds in the DNA minor groove—particularly in A–T–rich regions. This selective binding confers:
- High Signal-to-Noise in nuclear labeling
- Reliable DNA Quantitation based on fluorescence intensity
- Minimal Off-Target Background for clear imaging and flow cytometry
Why Fluorescence Increases Upon DNA Binding
In aqueous solution, free Hoechst 33342 exhibits a relatively low fluorescence quantum yield. However, once the dye inserts into the narrow A–T–rich minor groove:
In aqueous solution, free Hoechst 33342 exhibits a relatively low fluorescence quantum yield. However, once the dye inserts into the narrow A–T–rich minor groove:
- Structural Confinement: The dye’s molecular motion is restricted, reducing non-radiative energy loss.
- Water Exclusion: Fewer water molecules surround the fluorophore, minimizing quenching pathways.
- Hydrophobic Interactions: The ethyl substituent forms stabilizing contacts with consecutive A–T pairs, further enhancing fluorescence.
Together, these factors can boost Hoechst 33342 fluorescence by up to 20–30×, making it exceptionally bright once bound to DNA (Lakowicz, 2006).
Additional Utility and Specificity
- Minor Groove Affinity: Favors A–T–rich sequences, aiding specialized genomic or epigenetic studies (e.g., histone modification mapping).
- Low Off-Target Staining: Results in clean nuclear definition, particularly beneficial for high-content or multi-channel assays.
- Efflux Studies: Serves as a model substrate to investigate bacterial ABC transporters (e.g., BmrA), illuminating mechanisms of multidrug resistance (Di Cesare et al., 2024).
Researchers leverage these properties for a range of applications—from routine DNA quantification to probing DNA curvature and exploring conformational changes in complex assays.
Core Applications
Live-Cell and Fixed-Cell Nuclear Staining
- Live-Cell Versatility: Lipophilicity facilitates membrane penetration, enabling real-time monitoring of cell division and apoptosis—even in 3D spheroids or organoids.
- Fixed-Cell Complement: Hoechst 33342 provides a bright, stable nuclear counterstain in immunofluorescence protocols, synergizing well with red and green fluorophores.
Cell Cycle Analysis
- Flow Cytometry: Readily distinguishes G0/G1, S, and G2/M phases with minimal impact on cell viability.
- Sorting Applications: Can help isolate stem-like or progenitor cells based on dye efflux (Side Population analysis).
Apoptosis and Cell Death Detection
- Nuclear Morphology: Condensed or fragmented nuclei become distinctly visible, allowing clear discrimination of apoptotic cells.
- Viability Assays: Often combined with Annexin V or Propidium Iodide (PI) to differentiate between live, apoptotic, and necrotic populations.
High-Content Screening (HCS) and Automated Imaging
- Scalability: Ideal for automated nuclear segmentation at scale in multi-well plate formats.
- Low Background: Produces consistent, high-contrast signals, facilitating reliable readouts in drug screening.
Side Population (SP) and Stem Cell Analysis
- ABC Transporter Assays: Cells that actively efflux Hoechst 33342 form the "Side Population," aiding in identifying stem or progenitor cell fractions.
- Minimal Interference: Concentrations under 30 nM often minimize cytotoxic effects, enabling longer-term studies (Fuchs et al., 2023; Hu et al., 2024).
Additional Routine Applications
- Mycoplasma Detection: Reveals contamination as small, bright fluorescent spots (confirm via PCR).
- Super-Resolution & Multiplexing: Emits in the blue range, freeing other channels for additional probes.
Emerging Insights and Specialized Applications
Recent Literature Highlights
- Quiescent Cell Detection: Pairing Hoechst 33342 with Pyronin Y isolates rare G0 cells in leukemic co-cultures (Parker et al., 2024).
- Refined Phototoxicity Control: Reducing UV laser exposures (e.g., once every 30–60 minutes) limits bleaching and cell damage (Fuchs et al., 2023).
- Deep Learning Integration: Automated segmentation algorithms lower the need for additional immunofluorescent labels (Cooper, 2022).
Automated cell imaging [using Hoechst 33342] …include the ability to effectively compare adherent and suspension cell lines, reduced time to perform the assay, and environmental control allowing for long-term imaging studies. — Featherston et al., 2024
Experimental “Pearls”
- Concentration-Dependent Cytotoxicity: Recommended 5–30 nM range for extended live imaging (Fuchs et al., 2023).
- Light Exposure: Minimize high-intensity UV illumination to reduce photobleaching in multi-day protocols.
- Dual-/Triple-Staining with Hoechst 33342: Combine with Annexin V-FITC, PI, or TUNEL to distinguish apoptosis vs. necrosis.
- Buffer Considerations: pH 7.2–7.4 with ensures consistent fluorescence (Van den Berg van Saparoea et al., 2006).
Contrary to the prevailing assumption, Hoechst 33342 can be used in real-time imaging protocols for multiple days at sub-toxic concentrations, greatly expanding live-cell assay capacities. — Fuchs et al., 2023
Specialized & Emerging Applications
- Mitochondrial & Membrane Studies: Hoechst 33342 can be useful as an indicator in nano-thermometry or lipid-partitioning assays (Spicer, 2021; Cordeiro, 2023).
- Real-Time Gene Delivery Tracking: Sequential Hoechst additions for repeated viral transduction quantification (Hu et al., 2024).
- Microbial & Fungal Assays: Effective for visualizing protoplasts in pathogens like Phytophthora cinnamomi (Kharel et al., 2024).
Considerations for Specialized Applications
- pH and Ion Strength: Generally stable in physiological ranges; extreme pH shifts can reduce binding efficiency (Cordeiro, 2023).
- FRET/FLIM Potential: Hoechst 33342 can serve as a donor fluorophore in the blue range for advanced imaging techniques.
- Multiphoton Excitation: Near-infrared lasers help reduce phototoxicity in thicker samples (e.g., organoids).
- Partial DNA Saturation: Sub-saturating conditions allow ratiometric A–T analysis in certain genomic assays.
- High Autofluorescence Cell Lines: Titrate dye carefully; use gating strategies in flow cytometry to exclude debris.
- RNA/Mitochondrial DNA Labeling: Under specific conditions, Hoechst 33342 may bind RNA or mtDNA; maintaining low concentrations and short incubations usually prevents off-target staining.
- Photobleaching Strategies: Lower UV power and reduce exposure times to preserve signal.
- RBC Lysis: RBCs (no nuclei) remain unlabeled; remove or lyse RBCs in mixed populations for cleaner data.
- Membrane Transporter Assays: Hoechst 33342 often serves as a model substrate for bacterial ABC transporters (Hampton et al., 2024; Di Cesare et al., 2024), revealing real-time efflux and multidrug resistance profiles.
- Drug Resistance Reversal: Specialized membrane-fusing vehicles plus Hoechst 33342 help dissect transporter-inhibition strategies in cancer cells (Vahdati and Lamprecht, 2024).
By leveraging these detailed insights on Hoechst 33342—from concentration handling to advanced imaging approaches—researchers can optimize assays across a spectrum of cell biology, microbiology, and therapeutic studies.
Frequently Asked Questions
View All FAQ's
- What does Hoechst 33342 stain for?
Hoechst 33342 targets nuclear DNA, binding strongly in A–T–rich minor grooves. - Difference between Hoechst 33342 and DAPI?
Hoechst 33342 is more lipophilic, making it ideal for live-cell imaging, whereas DAPI is typically used on fixed or permeabilized cells. - Does Hoechst 33342 show cell death?
While not a classic viability dye, it does reveal nuclear fragmentation (e.g., in apoptotic cells). - Can I stain live cells with Hoechst 33342?
High membrane permeability makes Hoechst 33342 suited for live-cell applications. - What about cytotoxicity?
Hoechst 33342 can be cytotoxic at higher concentrations or extended exposures. We recommend sub-30 nM for multi-day imaging (Fuchs et al., 2023). - How long do Hoechst 33342-stained samples last?
Stained cells can retain fluorescence for days to weeks if kept protected from light at low temperatures. - Does Hoechst 33342 bind RNA or mitochondrial DNA?
Primarily binds double-stranded DNA. Incidental labeling of RNA or mitochondrial DNA (mtDNA) is minimal at standard concentrations. - Which is better, Hoechst 33342 or 33258?
Hoechst 33342 is often preferred for live cells; Hoechst 33258 typically suits fixed-cell setups. - How do I store Hoechst 33342 stock solution?
Keep at ≤−15 °C, shielded from light to maintain stability. - Does DAPI work for live cells?
DAPI is generally not recommended for live cells due to lower permeability and higher toxicity. - Any special concerns for multi-color imaging with Hoechst 33342?
Use a suitable filter set (~350 nm excitation, ~460 nm emission). Plan carefully if using green or red channels to avoid spectral crosstalk.
Further Reading
View All Citations
- Arvidsson, M., et al. “An Annotated High-Content Fluorescence Microscopy Dataset with Hoechst 33342-Stained Nuclei and Manually Labelled Outlines.”; Unpublished Dataset/Study, 2022.
- Cooper, J., et al. “Lymphocyte Classification from Hoechst-Stained Slides with Deep Learning.” 2022.
- Cordeiro, M. M., et al. “Interaction of Hoechst 33342 with POPC Membranes at Different pH Values.” 2023.
- Di Cesare, M., et al. (2024). “The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.” J Biol Chem, vol. 300, no. 1, p. 105546.
- Featherston, T., et al. (2024). “Comparing automated cell imaging with conventional methods of measuring cell proliferation and viability.” Toxicol Mech Methods, vol. 34, no. 8, pp. 886–896.
- Fuchs, H., et al. “Breaking a Dogma: High-Throughput Live-Cell Imaging in Real-Time with Hoechst 33342.” N.d., 2023. (Additional publication info not provided.)
- Gill, M. E., et al. “Isolation of Mouse Germ Cells by FACS Using Hoechst 33342 and SYTO16 Double Staining.” N.d., 2024. (Additional publication info not provided.)
- Goodell, M. A., et al. “Stem Cell Identification via Dye Efflux.” 1996, 1997.
- Hallap, T., et al. “Triple Fluorochrome Combination for Membrane Stability Testing.” 2006.
- Hampton, N., et al. (2024). “Strain-level variations of Dirofilaria immitis microfilariae in two biochemical assays.” PLoS One, vol. 19, no. 7, e0307261.
- Hou, Y., et al. “Salidroside Intensifies Mitochondrial Function...” 2023.
- Hu, X., et al. (2024). “Long-term in vitro monitoring of AAV-transduction efficiencies in real-time with Hoechst 33342.” PLoS One, vol. 19, no. 3, e0298173.
- Kharel, A., et al. (2024). “Viable protoplast isolation, organelle visualization and transformation of the globally distributed plant pathogen Phytophthora cinnamomi.” Protoplasma, vol. 261, no. 5, pp. 1073–1092.
- Kirby, J., et al. “The Dynamin Inhibitor...” 2024.
- Latt S. A., Wohlleb J. C. “Optical studies of the interaction of 33258 Hoechst with DNA, chromatin, and metaphase chromosomes.” Chromosoma. 1975-11-11; 52(4):297–316. doi: 10.1007/BF00364015. PMID: 1192901.
- Li, L., et al. “The DNA Minor-Groove Binding Agents Hoechst 33258 and 33342 Enhance Recombinant Adeno-Associated Virus (rAAV) Transgene Expression.” Journal of Gene Medicine, vol. 7, 2005, p. 420.
- Manzini, G., et al. “Nucleic Acids Research.” 1983.
- Merolli, A., et al. “Hoechst 33342 as a Marker for Imaging Neurites of Dorsal Root Ganglion in vitro.” N.d., 2022. (Additional journal info not provided.)
- Parker, J., et al. (2024). “Protocol for in vitro co-culture, proliferation, and cell-cycle analyses of patient-derived leukemia cells.” STAR Protoc, vol. 5, no. 3, p. 103202.
- Rahmé, R. “Assaying Cell-Cycle Status Using Flow Cytometry.” 2021.
- Rens, C., et al. “Apoptosis Assessment in High-Content and High-Throughput Screening Assays.” 2021.
- Spicer, G., et al. “Harnessing DNA for Nanothermometry.” 2021.
- Swain, B. M., et al. “Complexities of a Protonatable Substrate in Measurements of Hoechst 33342 Transport by Multidrug Transporter LmrP.” 2020.
- Takaoka, Y., et al. “Hoechst-Tagged Fluorescein Diacetate for the Fluorescence Imaging-Based Assessment of Stomatal Dynamics in Arabidopsis thaliana.” N.d., 2020. (Publication details not provided.)
- Vahdati, S., and Lamprecht, A. (2024). “Membrane-Fusing Vehicles for Re-Sensitizing Transporter-Mediated Multiple-Drug Resistance in Cancer.” Pharmaceutics, vol. 16, no. 4, p. 493.
- Van den Berg van Saparoea, B., et al. “Proton Motive Force-Dependent Hoechst 33342 Transport by the ABC Transporter LmrA of Lactococcus lactis.” Biochemistry, vol. 44, no. 1, 2006, pp. 1693–1700, https://doi.org/10.1021/bi051497y.
- Wang, F., et al. “Effective Detection of Hoechst Side-Population Cells by Flow Cytometry.” Journal of Visualized Experiments (JoVE), no. 210, 2024, e67012.
- Zhan, F., et al. “Minocycline Alleviates LPS-Induced Cognitive Dysfunction in Mice by Inhibiting the NLRP3/Caspase-1 Pathway.” Aging (Albany NY), vol. 16, no. 3, 2024, pp. 2989–3006.
- Zhang, X., and F. L. Kiechle. Annals of Clinical & Laboratory Science, 2006.
- Zheng, D., et al. (2024). “High-content image screening to identify chemical modulators for peroxisome and ferroptosis.” Cell Mol Biol Lett, vol. 29, no. 1, p. 26.
- Zhu, L., et al. “Schizandrin A Can Inhibit Non-Small Cell Lung Cancer Cell Proliferation...” 2021.
Calculators
Common stock solution preparation
Table 1. Volume of Water needed to reconstitute specific mass of Hoechst 33342 *Ultrapure Grade* to given concentration. Note that volume is only for preparing stock solution. Refer to sample experimental protocol for appropriate experimental/physiological buffers.0.1 mg | 0.5 mg | 1 mg | 5 mg | 10 mg | |
1 mM | 177.958 µL | 889.791 µL | 1.78 mL | 8.898 mL | 17.796 mL |
5 mM | 35.592 µL | 177.958 µL | 355.916 µL | 1.78 mL | 3.559 mL |
10 mM | 17.796 µL | 88.979 µL | 177.958 µL | 889.791 µL | 1.78 mL |
Molarity calculator
Enter any two values (mass, volume, concentration) to calculate the third.Citations
View all 38 citations: Citation Explorer
PD-L1 expression in cervical cancer tissue is strongly associated with the expression of CD73/TGF-$\beta$1, the percentage of CD8+/PD-1+ T cells and disease progression
Authors:
Ricardo, Mu{\~n}{\'o}z-God{\'\i}nez and Alberto, Monroy-Garc{\'\i}a and Mar{\'\i}a, Hern{\'a}ndez-Cueto {\'A}ngeles and Vadim, P{\'e}rez-Koldenkova and Gabriela, Molina-Castillo and de Lourdes, Mora-Garc{\'\i}a Mar{\'\i}a and others,
Journal:
Immunology Letters (2026): 107150
Efficient on-column removal of endotoxin from immunoglobulins such as AK23
Authors:
Rahimi, Siavash and Sauta, Patrizia and Edler, Monika and Locher, Elisabeth and Illi, Marlies and Shojaeian, Taravat and Borradori, Luca and Gentinetta, Thomas and Hariton, William VJ and M{\"u}ller, Eliane J
Journal:
Current Protocols (2025): e70238
VRK1 Is a Novel Therapeutic Target for Small Cell Neuroendocrine Carcinoma of the Cervix
Authors:
Kobayashi, Mariya and Nakagawa, Satoshi and Ishii, Yusuke and Kamei, Yuji and Kanda, Mizuki and Masuda, Tatsuo and Kakuda, Mamoru and Hiramatsu, Kosuke and Iwamiya, Tadashi and Egawa-Takata, Tomomi and others,
Journal:
Cancer Science (2025)
Cigarette smoke compromises macrophage innate sensing in response to pneumococcal infection
Authors:
Liao, Wei-Chih and Chou, Chia-Huei and Ho, Mao-Wang and Chen, Jo-Tsen and Chou, Shu-Ling and Huang, Mei-Zi and Bui, Ngoc-Niem and Wu, Hui-Yu and Lee, Chi-Fan and Huang, Wei-Chien and others,
Journal:
Journal of Microbiology, Immunology and Infection (2024)
LGP2 Facilitates Bacterial Escape through Binding Peptidoglycan via EEK Motif and Suppressing NOD2--RIP2 Axis in Cyprinidae and Xenocyprididae Families
Authors:
Liang, Bo and Li, Wenqian and Yang, Chunrong and Su, Jianguo
Journal:
The Journal of Immunology (2024)
References
View all 42 references: Citation Explorer
Usefulness of a triple fluorochrome combination Merocyanine 540/Yo-Pro 1/Hoechst 33342 in assessing membrane stability of viable frozen-thawed spermatozoa from Estonian Holstein AI bulls
Authors:
Hallap T, Nagy S, Jaakma U, Johannisson A, Rodriguez-Martinez H.
Journal:
Theriogenology (2006): 1122
Fatty acid synthase and its mRNA concentrations are decreased at different times following Hoechst 33342-induced apoptosis in BC3H-1 myocytes
Authors:
Zhang X, Kiechle FL.
Journal:
Ann Clin Lab Sci (2006): 185
The DNA minor groove binding agents Hoechst 33258 and 33342 enhance recombinant adeno-associated virus (rAAV) transgene expression
Authors:
Li L, Yang L, Kotin RM.
Journal:
J Gene Med (2005): 420
Resistance mechanism development to the topoisomerase-I inhibitor Hoechst 33342 by Leishmania donovani
Authors:
Marquis JF, Hardy I, Olivier M.
Journal:
Parasitology (2005): 197
Acid-base and electronic structure-dependent properties of Hoechst 33342
Authors:
Aleman C, Namba AM, Casanovas J.
Journal:
J Biomol Struct Dyn (2005): 29
Physical properties
| Molecular weight | 561.93 |
| Solvent | Water |
Spectral properties
| Excitation (nm) | 352 |
| Emission (nm) | 454 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 41116134 |
| CAS | 875756-97-1 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 350 nm |
| Emission | 461 nm |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 22, 2025

