Live or Dead™ Fixable Dead Cell Staining Kit
Blue Fluorescence
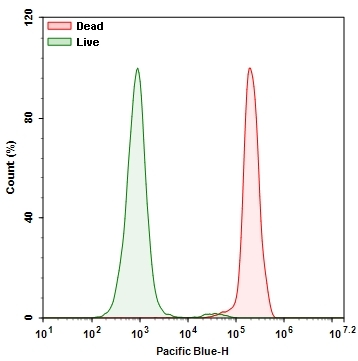
Our Live or Dead™ Fixable Dead Cell Staining Kits are a set of tools for labeling cells for fluorescence microscopic investigations of cell functions. The effective labeling of cells provides a powerful method for studying cellular events in a spatial and temporal context. This particular kit is designed to uniformly label fixed mammalian cells in blue fluorescence for long term microscopic examination. The kit uses a proprietary blue fluorescent dye that is more fluorescent upon bonding to cellular components. The fluorescent dye used in the kit is quite photostable so that the images can be repeatedly examined. The kit provides all the essential components with an optimized cell-labeling protocol. It is an excellent tool for preserving of fluorescent images of particular cells, and can also be used for fluorescence microscope demonstrations.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22600 | 200 Tests | Price |
Spectral properties
| Excitation (nm) | 346 |
| Emission (nm) | 434 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Flow cytometer | |
| Excitation | 355 nm or 405 nm laser |
| Emission | 450/40 nm filter |
| Instrument specification(s) | Pacific Blue channel |
| Fluorescence microscope | |
| Excitation | 353 nm |
| Emission | 442 nm |
| Recommended plate | Black wall/clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 17, 2026

