Portelite™ Fluorimetric DNA Quantitation Kit with Broad Dynamic Range
Optimized for Cytocite™ and Qubit™ Fluorometers
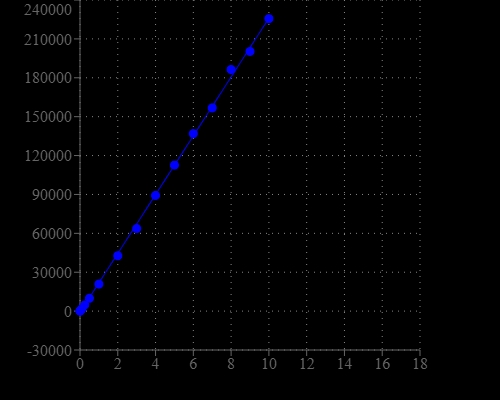
DNA Quantitation is a very important task in DNA sample preparations for various analyses. Portelite™ Fluorimetric DNA Quantitation Kit provides a rapid method to quantify dsDNA with Helixyte™ Green BR using a hand-held fluorometer. It is optimized for Cytocite™ and Qubit™ fluorometers. Portelite™ Fluorimetric DNA Quantitation assay is linear over three orders of magnitude. The assay is highly selective for double-stranded DNA (dsDNA) over RNA and is optimized for measuring dsDNA concentrations from 10 pg/µL to 10 ng/µL. Helixyte™ Green BR exhibits large fluorescence enhancement upon binding to dsDNA, and it is a few magnitudes more sensitive than UV absorbance readings.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17665 | 100 Tests | Price |
Storage, safety and handling
| Intended use | Research Use Only (RUO) |
Instrument settings
| Qubit Fluorometer | |
| Excitation | 480 nm |
| Emission | 530 nm |
| Instrument specification(s) | 0.2 mL PCR vial |
| CytoCite Fluorometer | |
| Excitation | 480 nm |
| Emission | 530 nm |
| Instrument specification(s) | 0.2 mL PCR vial |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 20, 2026
