Amplite® Colorimetric NADH Assay Kit
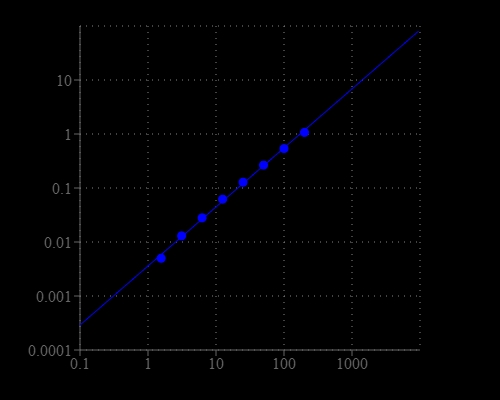
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) are two important cofactors found in cells. NADH is the reduced form of NAD+. NAD forms NADP with the addition of a phosphate group to the 2' position of the adenyl nucleotide through an ester linkage. The traditional NAD/NADH and NADP/NADPH assays are based on monitoring the changes in NADH or NADPH absorption at 340 nm. The short UV wavelength of NAD/NADH and NADP/NADPH assays makes the traditional methods suffer low sensitivity and high interference. Due to the weak absorption of NAD and NADH, the UV absorption method requires large sample sizes, making NAD and NADH measurements unpractical for limited sample size. AAT Bioquest's Amplite® Colorimetric NADH Assay Kit provides a convenient method for the detection of NADH. The NADH probe is a chromogenic sensor that has its maximum absorbance at 460 nm upon NADH reduction. The absorbance increase at 460 nm is directly proportional to the concentration of NADH in the solution. The NADH probe can recognize NADH in an enzyme-free reaction, and the signal can be easily read by an absorbance microplate reader at ~460 nm. The Amplite® Colorimetric NADH Assay Kit provides a sensitive assay to detect as little as 3 µM NADH in a 100 µL assay volume. The assay can be performed in a convenient 96-well or 384-well microtiter-plate format.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 15271 | 400 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Absorbance microplate reader | |
| Absorbance | 460 nm |
| Recommended plate | Clear bottom |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 24, 2026
