DAPI
4,6-Diamidino-2-phenylindole, dihydrochloride; CAS 28718-90-3
DAPI (4',6-diamidino-2-phenylindole) is a cell-impermeant blue fluorescent DNA stain that binds to AT-rich regions in the minor groove, universally used for nuclear counterstaining in fixed cells and identifying dead cells in live samples.
- AT-Rich Minor Groove Binder: 20-fold fluorescence enhancement upon dsDNA binding (Kd ~100 nM)
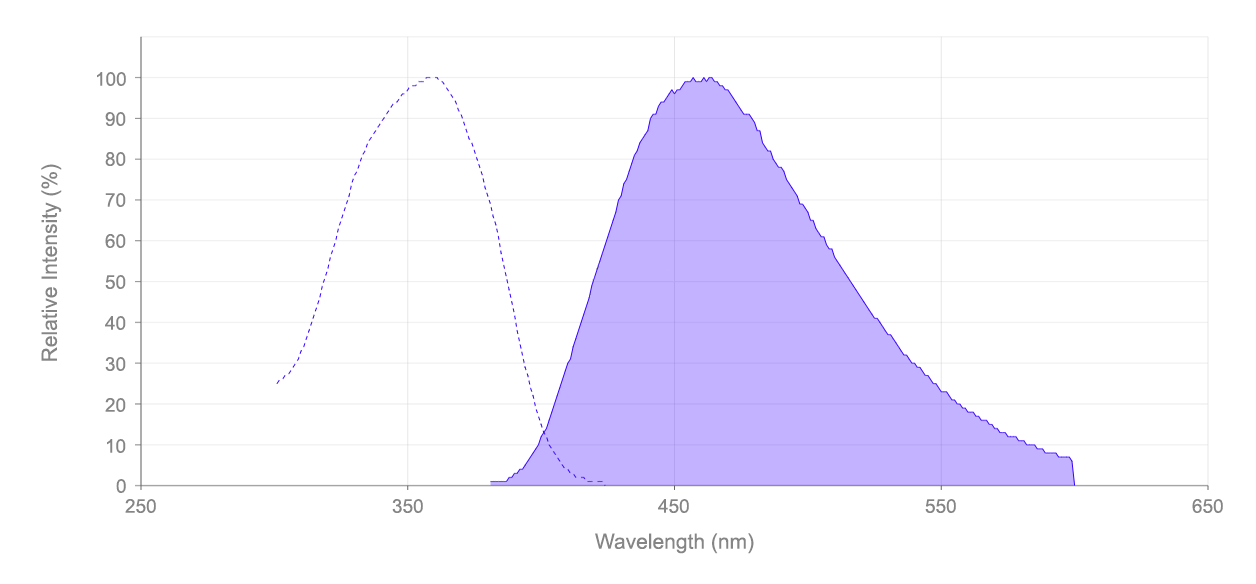
- UV Excitation/Blue Emission: Ex/Em = 358/461 nm, compatible with DAPI filter sets
- Nuclear Counterstain Standard: Cell-impermeant for fixed cell nuclear visualization
- Dead Cell Indicator: Membrane impermeability restricts staining to compromised cells in live samples

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17510 | 10 mg | Price | |
| 17511 | 100 mg | Price | |
| 17513 | 25 mg | Price |
Physical properties
| Molecular weight | 350.25 |
| Solvent | Water |
Spectral properties
| Extinction coefficient (cm -1 M -1) | 27000 |
| Excitation (nm) | 359 |
| Emission (nm) | 457 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 41116134 |
| CAS | 28718-90-3 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026

