Helixyte™ Green ssDNA reagent
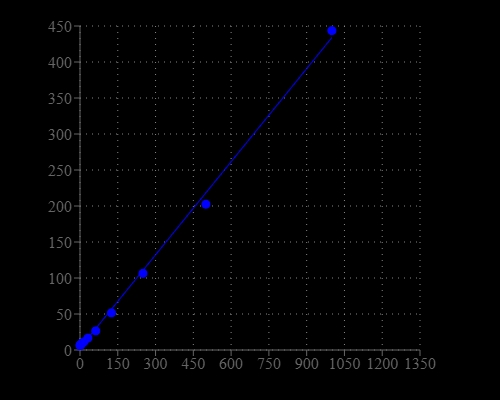
Synthetic oligonucleotides are used in several molecular biology techniques, such as DNA sequencing, site-directed mutagenesis, DNA amplification, and in situ hybridization. The most commonly used technique for measuring oligonucleotide and single-stranded DNA (ssDNA) concentration is the determination of absorbance at 260 nm (A260). However, absorbance methods suffer significant interferences resulting from various contaminants commonly found in nucleic acid preparations, including nucleotides, double-stranded nucleic acids, and proteins. Helixyte™ Green ssDNA Quantifying Reagent is an excellent alternative for quantifying ssDNA and oligonucleotides with significantly improved sensitivity and selectivity. It is a positively charged fluorescent probe that binds to the hydrophobic pockets of ssDNA and forms a highly luminescent complex through the synergistic actions of stacking, hydrophobic forces, hydrogen bonding, and electrostatic interactions. Helixyte™ Green ssDNA reagent has extremely low inherent fluorescence that is significantly enhanced upon binding to ssDNA. The increased sensitivity of Helixyte™ Green ssDNA over absorbance at 260 nm enables researchers to quantify as little as 100 pg/mL oligonucleotide or ssDNA (~500 pg/mL) with a standard spectrofluorometer and fluorescein excitation and emission wavelengths. As little as 1 ng/mL oligonucleotide or ssDNA can be detected with a fluorescence microplate reader. Helixyte™ Green ssDNA reagent has a wide linear range of detection ranging from 100 pg/mL to 1 μg/mL.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17620 | 1000 Tests | Price | |
| 17621 | 10000 Tests | Price |
Physical properties
| Solvent | DMSO |
Spectral properties
| Excitation (nm) | 498 |
| Emission (nm) | 519 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 28, 2026

