Screen Quest™ Fluorimetric MDR Assay Kit
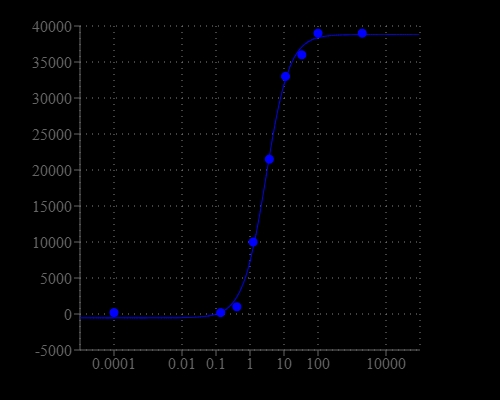
Tumor cell resistance to cytotoxic drugs is considered one of the major obstacles to successful chemotherapy. Some tumors are initially resistant and never respond to cytostatic drug treatment; others initially respond well but eventually regrow and become resistant. This phenomenon may result from genetic mutations induced by the administered antitumor agent, or may represent the selection of preexisting resistant cell populations in the malignant tumor. Multi-drug resistance (MDR) is a major factor in the failure of many forms of chemotherapy. In the past few years it has become widely accepted that the resistance to chemotherapy correlates with the overexpression of at least two ATP-dependent drug-efflux pumps. These cell membrane proteins, called P-glycoprotein (Pgp, MDR1), and multidrug-resistance-associated protein (MRP1) are members of the ABC transporter family. Our assay kit uses a fluorescent MDR indicator for assaying these two MDR pump activities. This hydrophobic fluorescent dye molecule rapidly penetrates cell membranes and becomes trapped in cells. Following a short incubation, the intracellular free dye concentration can increase significantly. In the MDR1 and/or MRP1-expressing cells this dye is extruded by the MDR transporter, thus decreasing the cellular fluorescence intensity. However, when its extrusion is blocked by an agent that interferes with the MDR1 and/or MRP1 pump-activity, its cellular fluorescence intensity increases significantly. Our MDR assay kit provides all the essential components with an optimized assay method. The assay can be performed in a convenient 96-well or 384-well microtiter-plate format and easily adapted to automation. This assay kit is ideal for high throughput screening of MDR pump inhibitors or identifying the cells that have high level of MDR pump activities.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 36340 | 100 Tests | Price | |
| 36341 | 1000 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 7, 2026
