Cell Meter™ NIR Mitochondrion Membrane Potential Assay Kit
Optimized for Microplate Reader
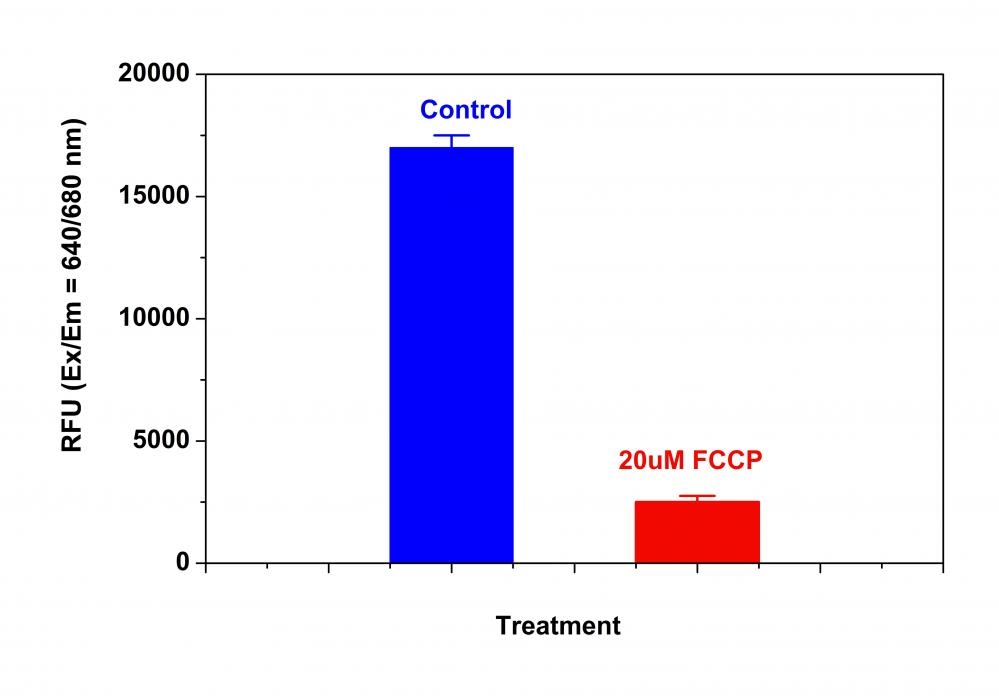
Our Cell Meter™ assay kits are a set of tools for monitoring cell viability. There are a variety of parameters that can be used for monitoring cell viability. This particular kit is designed to monitor cell apoptosis through measuring the loss of the mitochondrial membrane potential. The collapse of mitochondrial membrane potential coincides with the opening of the mitochondrial permeability transition pores, leading to the release of cytochrome C into the cytosol, which in turn triggers other downstream events in the apoptotic cascade. Our Cell Meter™ NIR Mitochondria Membrane Potential Detection Kit provides all the essential components with an optimized assay method for the detection of apoptosis in cells with the loss of mitochondrial membrane potential. This fluorometric assay is based on the detection of the mitochondrial membrane potential in cells by our proprietary cationic MitoLite NIR™ dye. In normal cells, MitoLite NIR™ accumulates primarily in mitochondria, however, in apoptotic cells, MitoLite NIR™ staining intensity decreases. Cells stained with MitoLite NIR™ can be monitored fluorimetrically at 660-680 nm with excitation of 620-640 nm. The kit can be used for screening of apoptosis activators and inhibitors. The assay can be performed in a convenient 96-well and 384-well fluorescence microtiter-plate format.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22803 | 500 Tests | Price |
Spectral properties
| Excitation (nm) | 640 |
| Emission (nm) | 657 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 640 nm |
| Emission | 680 nm |
| Cutoff | 665 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Bottom read mode |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 19, 2026

