StrandBrite™ Green Fluorimetric RNA Quantitation Kit
High Selectivity
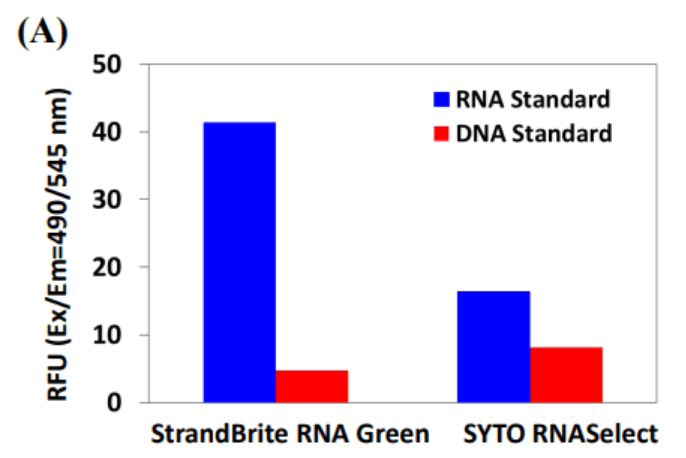
The major challenge to analyze RNA in live cells is the interferences caused by DNA. To address these difficulties, AAT Bioquest has developed the StrandBrite™ RNA Green, an excellent RNA-selective probe that generates significantly enhanced green fluorescence upon binding to RNA. It has been successfully used for flow cytometric analysis of live cells. StrandBrite™ RNA Green readily gets into live cells. It has the excitation/emission of 490/540 nm. In the DNase digest test, no significant change of fluorescence intensity in fixed cells stained with StrandBrite RNA Green was observed. In contrast, after RNase digestion, the initial fluorescence signal decreased immediately. These results indicate that initial fluorescence signal was generated from the specific interaction of StrandBrite RNA Green with RNA in cells. Short exposure of live cells to antinomycin D did cause inhibition of RNA synthesis during 6 hours after drug removal in a dose-dependent manner. These data demonstrate that StrandBrite RNA Green is a sensitive RNA-selective dye for staining nucleolar RNA in live and fixed cells. StrandBrite RNA Green has less DNA interferences than the commonly used SYTO® RNASelect™ dye. StrandBrite™ RNA Green is a highly RNA-selective fluorescent probe. Due to its excellent cell permeability and spectral properties, it has been successfully used for flow cytometric RNA analysis and fluorescence microscope in live cells. It can be well excited with the 488 nm blue laser and monitored in FITC channel. StrandBrite™ RNA Green provides a valuable method for identifying and labeling cells with a single incubation step and can discriminate RNA from DNA with better selectivity than the commonly used SYTO® RNASelect™.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17657 | 100 Tests | Price |
Spectral properties
| Excitation (nm) | 509 |
| Emission (nm) | 527 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| UNSPSC | 41116134 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 540 nm |
| Cutoff | 515 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 31, 2026

