Helixyte™ Gold Nucleic Acid Gel Stain
10,000X DMSO Solution
Helixyte™ Gold is a highly sensitive nucleic acid gel stain (equivalent to SYBR Gold) with broad specificity for DNA and RNA that provides superior detection of nucleic acids in agarose and polyacrylamide gels.
- Industry Standard Compatibility: Equivalent performance to detect nucleic acids in agarose and polyacrylamide gels, with a strong preference for DNA over RNA
- Reliable Performance: Proven effectiveness across multiple experimental conditions
- Versatile Applications: Suitable for a wide range of research and diagnostic applications

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17595 | 1 ml | Price |
Physical properties
| Molecular weight | 749.48 |
| Solvent | DMSO |
Spectral properties
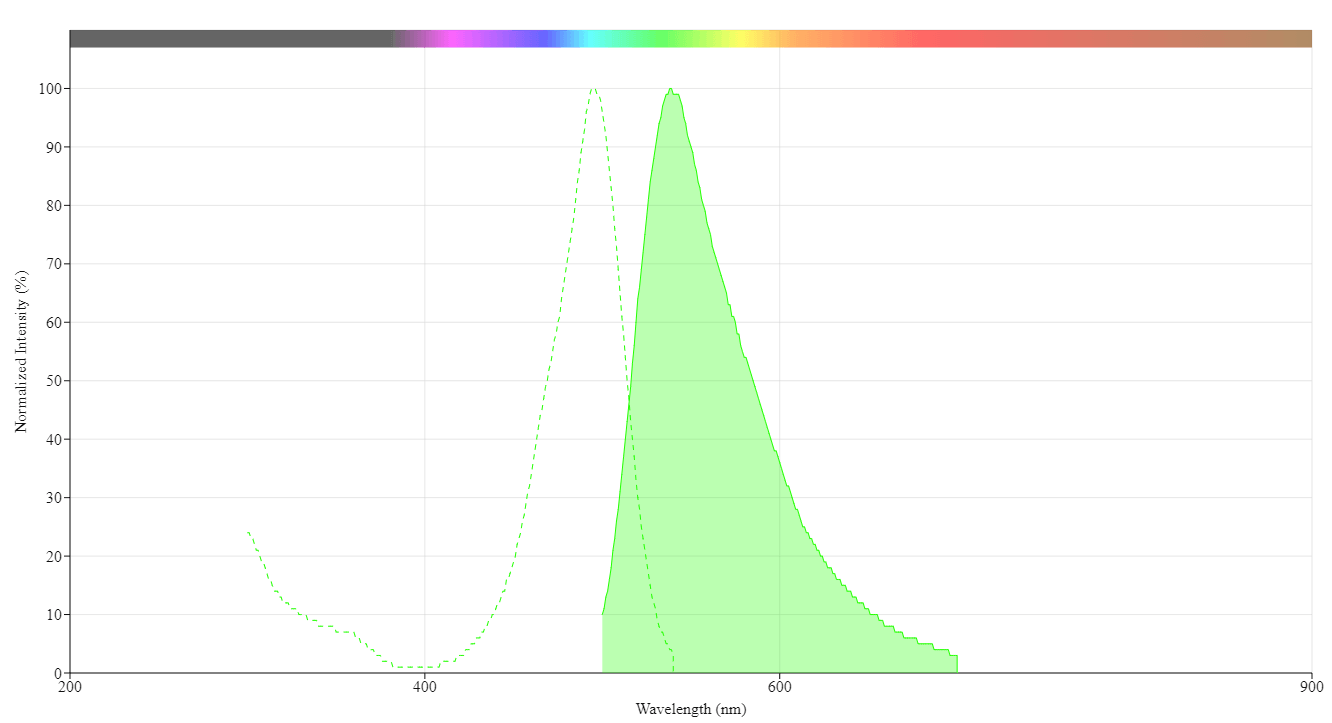
| Excitation (nm) | 496 |
| Emission (nm) | 539 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 41116134 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on September 23, 2024

