MycoLight™ Fluorescence Live/Dead Bacterial Imaging Kit
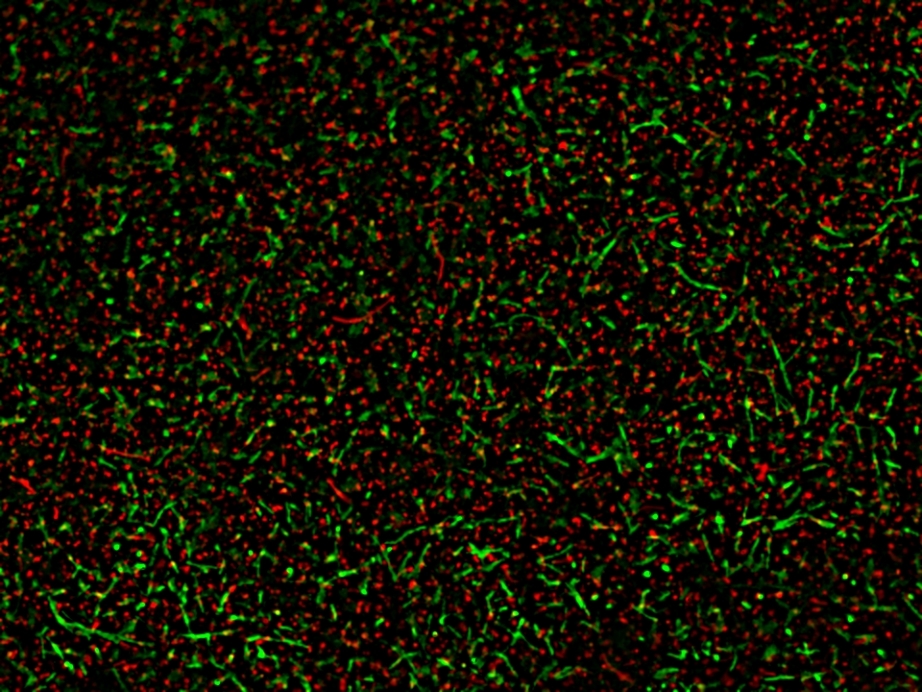
AAT Bioquest's MycoLight™ Fluorescence Live/Dead Bacterial Imaging Kit provides two-color fluorescence assay for visualizing live and dead bacteria through fluorescent microscope. MycoLight™ 520 is a non-fluorescent esterase substrate that diffuses into both Gram positive and Gram-negative bacteria. Upon hydrolysis by bacterial intracellular non-specific esterases, a green fluorescent product is produced and accumulated within bacteria. In contrast, propidium iodide is a red-fluorescent nucleic acid stain that only penetrates bacteria with damaged membranes. Thus, with an appropriate mixture of the MycoLight™ 520 and propidium iodide stains, live bacteria with intact cell membranes emits green fluorescence, whereas dead bacteria with damaged membranes gives red fluorescence. The MycoLight™ Fluorescence Live/Dead Bacterial Imaging Kit is a robust tool for imaging Live/Dead bacteria. Stained cells can be monitored fluorimeterically (FITC filter set) and (TRITC filter set) for live and dead bacteria respectively.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22411 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 488 nm / 540 nm |
| Emission | 530 nm / 620 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | FITC / TRITC filter(s) |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on January 30, 2026
