MycoLight™ Rapid Fluorescence Gram-Positive Bacteria Staining Kit
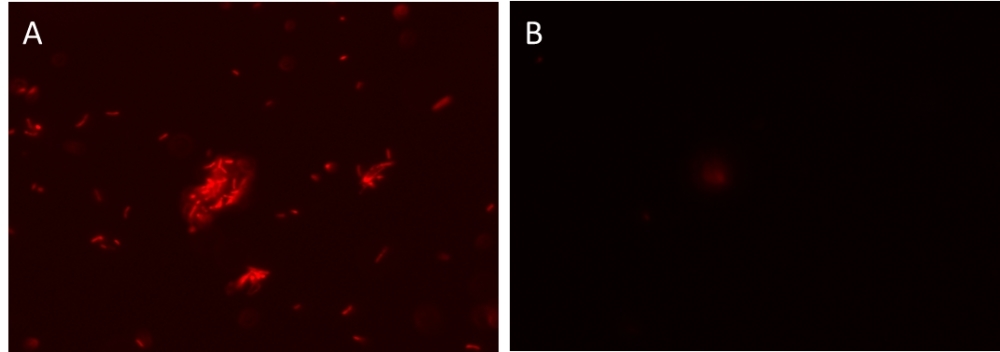
AAT Bioquest's MycoLight™ Rapid Fluorescence Gram-Positive Bacteria Staining Kit provides a novel one-step fluorescence assay for the determination of gram sign in living bacteria. The gram stain is an important and widely used method for the taxonomic classification of bacteria in clinical and research settings. The original method involves quite a few steps like heat fixation, two-steps staining protocol, alcohol extraction and counterstaining. These steps can create inconsistent staining. AAT Bioquest's one-step kit overcomes the existing problems by eliminating the labor-intensive steps. The kit uses a fluorescently labeled Concanavalin A (ConA), which is a lectin that selectively binds to N-acetyl glucosamine exposed on the surface of gram-positive bacteria. When gram-negative and gram-positive bacteria are stained with the fluorescently labeled ConA conjugate, only gram-positive bacteria fluoresce red. Stained bacteria can be monitored fluorimeterically. Our kit is robust and convenient since the fluorescently labeled ConA conjugate used in our kit demonstrates higher brightness and photo stability over other existing dyes.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22415 | 100 Tests | Price |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | 650 nm |
| Emission | 669 nm |
| Recommended plate | Black wall/clear bottom |
| Instrument specification(s) | Cy5 filter |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 10, 2026
