ReadiUse™ bacterial cell lysis buffer
5X
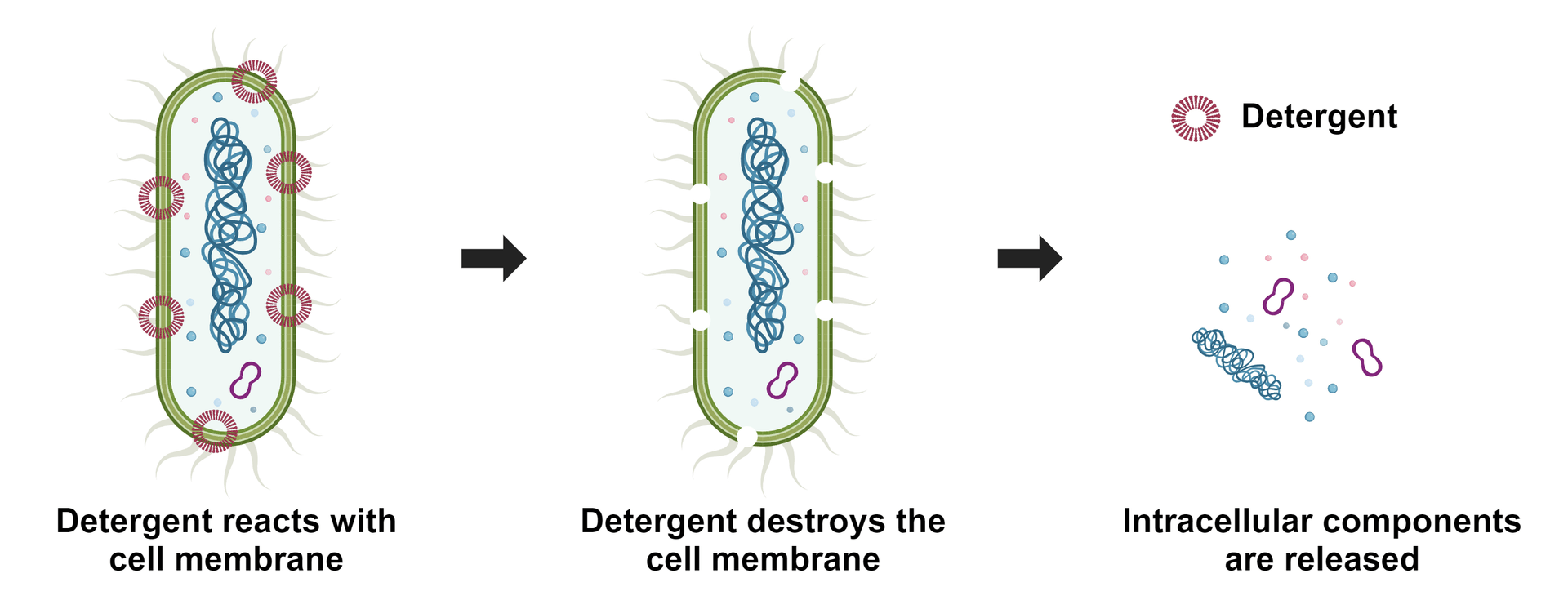
Cell lysis refers to the breaking down of cells, and it is often used to analyze specific cellular compositions such as proteins, lipids, nucleic acids, reporter molecules, cell signal molecules and other small biomolecules. Depending upon the detergents used, either all or some membranes are lysed. ReadiUse™ reagents require minimal hands-on time. This bacterial cell lysis buffer just requires a simple 5-fold dilution. It is widely used for lyzing cells for quantifying small biological molecules such as NAD(P)/NAD(P)H measurement in bacteria.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 24100 | 10 mL | Price |
Storage, safety and handling
| Intended use | Research Use Only (RUO) |
| Storage | Refrigerated (2-8 °C) |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 7, 2026
