ReadiCleave™ XFD488 AML-NHS ester
Ordering information
| Price | |
| Catalog Number | |
| Unit Size | |
| Quantity |
Additional ordering information
| Telephone | 1-800-990-8053 |
| Fax | 1-800-609-2943 |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Shipping | Standard overnight for United States, inquire for international |
Physical properties
| Molecular weight | 1340.49 |
| Solvent | DMSO |
Spectral properties
| Correction Factor (260 nm) | 0.3 |
| Correction Factor (280 nm) | 0.11 |
| Extinction coefficient (cm -1 M -1) | 73000 |
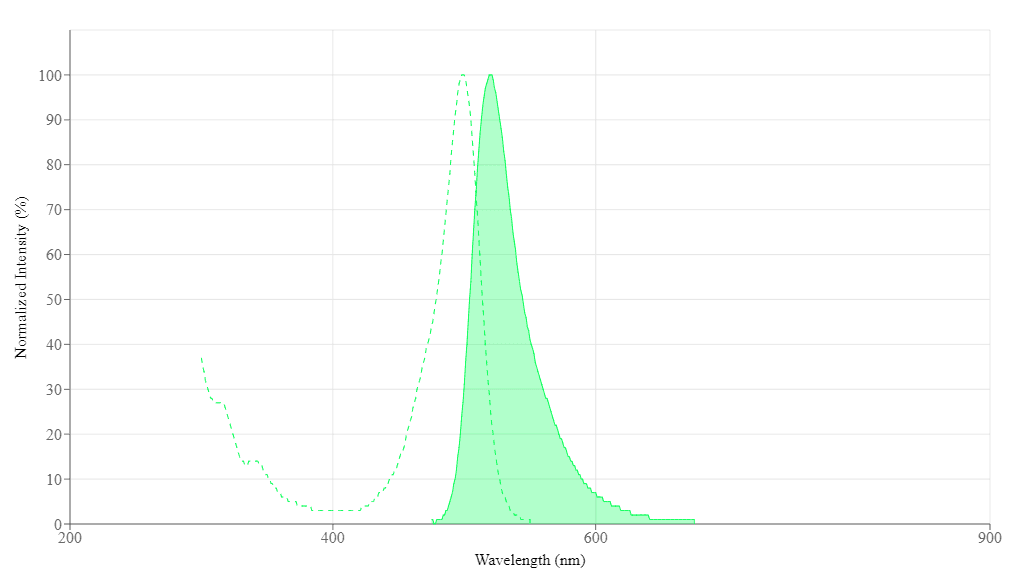
| Excitation (nm) | 499 |
| Emission (nm) | 520 |
| Quantum yield | 0.921 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
Direct upgrades
| ReadiCleave™ iFluor 488 AML-NHS ester |
Related products
| Overview |
Molecular weight 1340.49 | Correction Factor (260 nm) 0.3 | Correction Factor (280 nm) 0.11 | Extinction coefficient (cm -1 M -1) 73000 | Excitation (nm) 499 | Emission (nm) 520 | Quantum yield 0.921 |
Fluorescence-based methods have many advantages for biological detections in terms of sensitivity and convenience. Many biological molecules can be readily labeled with a fluorescent tag for fluorescence imaging and flow cytometry analysis. However, most of the existing fluorescent tags are used to permanently labeling biological targets from which the added fluorescent tags cannot be cleaved for further downstream analysis, such as mass spectral analysis. AAT Bioquest’s ReadiCleave™ linkers enable fluorescent tags conjugated to a biological target from which the added fluorescent tag can be removed when needed. This ReadiCleave™ AML XFD488 contains an azidomethyl linker that can be cleaved with TCEP to remove the Alexa Fluor® 488 fluorophore from the target molecule. The cleavage can be carried out by adding 10 mM TCEP solution (pH 7.5), and incubating at 65 °C for 1-5 min. XFD488 is the same fluorophore to Alexa Fluor® 488 (Alexa Fluor® is the trademark of ThermoFisher).
Calculators
Common stock solution preparation
Table 1. Volume of DMSO needed to reconstitute specific mass of ReadiCleave™ XFD488 AML-NHS ester to given concentration. Note that volume is only for preparing stock solution. Refer to sample experimental protocol for appropriate experimental/physiological buffers.
| 0.1 mg | 0.5 mg | 1 mg | 5 mg | 10 mg | |
| 1 mM | 74.6 µL | 372.998 µL | 745.996 µL | 3.73 mL | 7.46 mL |
| 5 mM | 14.92 µL | 74.6 µL | 149.199 µL | 745.996 µL | 1.492 mL |
| 10 mM | 7.46 µL | 37.3 µL | 74.6 µL | 372.998 µL | 745.996 µL |
Molarity calculator
Enter any two values (mass, volume, concentration) to calculate the third.
| Mass (Calculate) | Molecular weight | Volume (Calculate) | Concentration (Calculate) | Moles | ||||
| / | = | x | = |
Spectrum
Open in Advanced Spectrum Viewer

Spectral properties
| Correction Factor (260 nm) | 0.3 |
| Correction Factor (280 nm) | 0.11 |
| Extinction coefficient (cm -1 M -1) | 73000 |
| Excitation (nm) | 499 |
| Emission (nm) | 520 |
| Quantum yield | 0.921 |
Product Family
| Name | Excitation (nm) | Emission (nm) | Extinction coefficient (cm -1 M -1) | Quantum yield | Correction Factor (260 nm) | Correction Factor (280 nm) |
| ReadiCleave™ AML Cy5 NHS ester | 651 | 670 | 2500001 | 0.271, 0.42 | 0.02 | 0.03 |
| ReadiCleave™ XFD532 AML-NHS ester | 534 | 553 | 81000 | 0.611 | 0.24 | 0.09 |
| ReadiCleave™ FITC AML-NHS ester | 498 | 517 | 800001 | 0.79001, 0.952 | 0.32 | 0.35 |
| ReadiCleave™ XFD546 AML-NHS ester | 561 | 572 | 112000 | 0.791 | 0.21 | 0.12 |
| ReadiCleave™ XFD647 AML-NHS ester | 650 | 671 | 239000 | 0.331 | 0.00 | 0.03 |
References
View all 50 references: Citation Explorer
Alexa Fluor 488-conjugated cholera toxin subunit B optimally labels neurons 3-7 days after injection into the rat gastrocnemius muscle.
Authors: Cui, Jing-Jing and Wang, Jia and Xu, Dong-Sheng and Wu, Shuang and Guo, Ya-Ting and Su, Yu-Xin and Liu, Yi-Han and Wang, Yu-Qing and Jing, Xiang-Hong and Bai, Wan-Zhu
Journal: Neural regeneration research (2022): 2316-2320
Authors: Cui, Jing-Jing and Wang, Jia and Xu, Dong-Sheng and Wu, Shuang and Guo, Ya-Ting and Su, Yu-Xin and Liu, Yi-Han and Wang, Yu-Qing and Jing, Xiang-Hong and Bai, Wan-Zhu
Journal: Neural regeneration research (2022): 2316-2320
Detection of Homologous Blood Transfusion in Sport Doping by Flow Cytofluorimetry: State of the Art and New Approaches to Reduce the Risk of False-Negative Results.
Authors: Donati, Francesco and de la Torre, Xavier and Pagliarosi, Sarajane and Pirri, Daniela and Prevete, Giuliana and Botrè, Francesco
Journal: Frontiers in sports and active living (2022): 808449
Authors: Donati, Francesco and de la Torre, Xavier and Pagliarosi, Sarajane and Pirri, Daniela and Prevete, Giuliana and Botrè, Francesco
Journal: Frontiers in sports and active living (2022): 808449
A fluorescent aptasensor for the detection of Aflatoxin B1 by graphene oxide mediated quenching and release of fluorescence.
Authors: Setlem, Sai Keerthana and Mondal, Bhairab and Ramlal, Shylaja
Journal: Journal of microbiological methods (2022): 106414
Authors: Setlem, Sai Keerthana and Mondal, Bhairab and Ramlal, Shylaja
Journal: Journal of microbiological methods (2022): 106414
Introducing Cysteines into Nanobodies for Site-Specific Labeling.
Authors: Hansen, Simon Boje and Andersen, Kasper Røjkjær
Journal: Methods in molecular biology (Clifton, N.J.) (2022): 327-343
Authors: Hansen, Simon Boje and Andersen, Kasper Røjkjær
Journal: Methods in molecular biology (Clifton, N.J.) (2022): 327-343
Dynamics of intracellular neonatal Fc receptor-ligand interactions in primary macrophages using biophysical fluorescence techniques.
Authors: Pannek, Andreas and Houghton, Fiona J and Verhagen, Anne M and Dower, Steven K and Hinde, Elizabeth and Gleeson, Paul A
Journal: Molecular biology of the cell (2022): ar6
Authors: Pannek, Andreas and Houghton, Fiona J and Verhagen, Anne M and Dower, Steven K and Hinde, Elizabeth and Gleeson, Paul A
Journal: Molecular biology of the cell (2022): ar6
[The Role of Zyxin in Regulating Platelet Cytoskeleton Distribution].
Authors: Cheng, Bin and Yan, Rong and Zhang, Su-Qin and Yang, Meng-Nan and Dai, Ke-Sheng
Journal: Zhongguo shi yan xue ye xue za zhi (2021): 876-880
Authors: Cheng, Bin and Yan, Rong and Zhang, Su-Qin and Yang, Meng-Nan and Dai, Ke-Sheng
Journal: Zhongguo shi yan xue ye xue za zhi (2021): 876-880
Cholecystokinin-B Receptor-Targeted Nanoparticle for Imaging and Detection of Precancerous Lesions in the Pancreas.
Authors: Smith, Jill P and Cao, Hong and Edmondson, Elijah F and Dasa, Siva Sai Krishna and Stern, Stephan T
Journal: Biomolecules (2021)
Authors: Smith, Jill P and Cao, Hong and Edmondson, Elijah F and Dasa, Siva Sai Krishna and Stern, Stephan T
Journal: Biomolecules (2021)
Broad immunophenotyping of the murine brain tumor microenvironment.
Authors: Tomaszewski, W H and Waibl-Polania, J and Miggelbrink, A M and Chakraborty, M A and Fecci, P E and Sampson, J H and Gunn, M D
Journal: Journal of immunological methods (2021): 113158
Authors: Tomaszewski, W H and Waibl-Polania, J and Miggelbrink, A M and Chakraborty, M A and Fecci, P E and Sampson, J H and Gunn, M D
Journal: Journal of immunological methods (2021): 113158
Polarization Angle Dependence of Optical Gain in a Hybrid Structure of Alexa-Flour 488/M13 Bacteriophage.
Authors: Kim, Inhong and Jang, Juyeong and Lee, Seunghwan and Kim, Won-Geun and Oh, Jin-Woo and Wang, Irène and Vial, Jean-Claude and Kyhm, Kwangseuk
Journal: Nanomaterials (Basel, Switzerland) (2021)
Authors: Kim, Inhong and Jang, Juyeong and Lee, Seunghwan and Kim, Won-Geun and Oh, Jin-Woo and Wang, Irène and Vial, Jean-Claude and Kyhm, Kwangseuk
Journal: Nanomaterials (Basel, Switzerland) (2021)
Cytoskeletal Actin Structure in Osteosarcoma Cells Determines Metastatic Phenotype via Regulating Cell Stiffness, Migration, and Transmigration.
Authors: Kita, Kouji and Asanuma, Kunihiro and Okamoto, Takayuki and Kawamoto, Eiji and Nakamura, Koichi and Hagi, Tomohito and Nakamura, Tomoki and Shimaoka, Motomu and Sudo, Akihiro
Journal: Current issues in molecular biology (2021): 1255-1266
Authors: Kita, Kouji and Asanuma, Kunihiro and Okamoto, Takayuki and Kawamoto, Eiji and Nakamura, Koichi and Hagi, Tomohito and Nakamura, Tomoki and Shimaoka, Motomu and Sudo, Akihiro
Journal: Current issues in molecular biology (2021): 1255-1266
Application notes
A New Protein Crosslinking Method for Labeling and Modifying Antibodies
Abbreviation of Common Chemical Compounds Related to Peptides
Bright Tide Fluor™-Based Fluorescent Peptides and Their Applications In Drug Discovery and Disease Diagnosis
FITC (Fluorescein isothiocyanate)
Fluorescein isothiocyanate (FITC)
Abbreviation of Common Chemical Compounds Related to Peptides
Bright Tide Fluor™-Based Fluorescent Peptides and Their Applications In Drug Discovery and Disease Diagnosis
FITC (Fluorescein isothiocyanate)
Fluorescein isothiocyanate (FITC)
