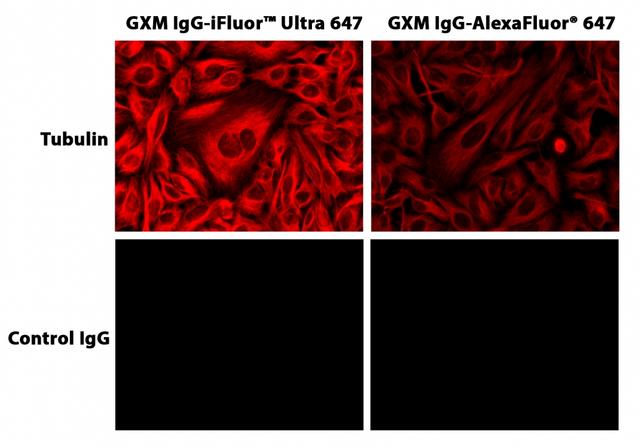
iFluor® Ultra 647 succinimidyl ester
Example protocol
PREPARATION OF STOCK SOLUTIONS
Unless otherwise noted, all unused stock solutions should be divided into single-use aliquots and stored at -20 °C after preparation. Avoid repeated freeze-thaw cycles
Mix 100 µL of a reaction buffer (e.g., 1 M sodium carbonate solution or 1 M phosphate buffer with pH ~8.5) with 900 µL of the target protein solution (e.g. antibody, protein concentration >2 mg/mL if possible) to give 1 mL protein labeling stock solution.
Note: The pH of the protein solution (Solution A) should be 8.5 ± 0.5. If the pH of the protein solution is lower than 8.0, adjust the pH to the range of 8.0-9.0 using 1 M sodium bicarbonate solution or 1 M pH 9.0 phosphate buffer.
Note: The protein should be dissolved in 1X phosphate buffered saline (PBS), pH 7.2-7.4. If the protein is dissolved in Tris or glycine buffer, it must be dialyzed against 1X PBS, pH 7.2-7.4, to remove free amines or ammonium salts (such as ammonium sulfate and ammonium acetate) that are widely used for protein precipitation.
Note: Impure antibodies or antibodies stabilized with bovine serum albumin (BSA) or gelatin will not be labeled well. The presence of sodium azide or thimerosal might also interfere with the conjugation reaction. Sodium azide or thimerosal can be removed by dialysis or spin column for optimal labeling results.
Note: The conjugation efficiency is significantly reduced if the protein concentration is less than 2 mg/mL. For optimal labeling efficiency the final protein concentration range of 2-10 mg/mL is recommended.
Add 100 µL high-quality, anhydrous dimethylsulfoxide (DMSO) or dimethyl-formamide (DMF) to 1 mg iFluor™ Ultra 647 SE to prepare 10 mg/mL stock solution. Mix well by pipetting or vortex.
Note: Prepare the dye stock solution (Solution B) before starting the conjugation. Use promptly. Extended storage of the dye stock solution may reduce the dye activity. Solution B can be stored in freezer for two weeks when kept from light and moisture. Avoid freeze-thaw cycles.
Note: Once reconstituted, the NHS ester reactive dye solution is not very stable, especially if exposed to moisture. It could hydrolyze into the nonreactive free acid in aqueous solutions.
SAMPLE EXPERIMENTAL PROTOCOL
This labeling protocol was developed for the conjugate of Goat anti-mouse IgG with iFluor™ Ultra 647 SE. You might need further optimization for your particular proteins.
Note: Each protein requires distinct dye/protein ratio, which also depends on the properties of dyes. Over labeling of a protein could detrimentally affects its binding affinity while the protein conjugates of low dye/protein ratio gives reduced sensitivity.
Use 6-10 molar ratio of Solution B (dye)/Solution A (protein) as the starting point: Add 5 µL of the dye stock solution (Solution B, assuming the dye stock solution is 10 mM) into the vial of the protein solution (95 µL of Solution A) with effective shaking.
Note: We recommend to use 10:1 molar ratio of Solution B (dye)/Solution A (protein). If it is too less or too high, determine the optimal dye/protein ratio at 5:1, 15:1 and 20:1 respectively.
- Continue to rotate or shake the reaction mixture at room temperature for 30-60 minutes.
The following protocol is an example of dye-protein conjugate purification with 15 mL MWCO=30K filter (https://www.emdmillipore.com/US/en/product/Amicon-Ultra-4-Centrifugal-Filter-Units,MM_NF-C7719)
- Prepare column according to the manufacture instruction.
- Load the reaction mixture (From "Run conjugation reaction") to the top of the column.
- Add 4 mL of PBS (pH 7.2-7.4) as soon as the sample runs.
- Centrifuge to concentrate to ~0.4 mL.
- Add 4 mL PBS and then concentrate to ~0.4 mL.
- Repeat ~3 times, until the elution absorbance at 650 nm < 0.1.
- Collect the purified Antibody-iFluor™Ultra 647 conjugate solution.
Characterize the Desired Dye-Protein Conjugate
The Degree of Substitution (DOS) is the most important factor for characterizing dye-labeled protein. Proteins of lower DOS usually have weaker fluorescence intensity, but proteins of higher DOS (e.g. DOS > 6) tend to have reduced fluorescence too. The optimal DOS for most antibodies is recommended between 2 and 10 depending on the properties of dye and protein. For effective labeling, the degree of substitution should be controlled to have 6-8 moles of iFluor™ Ultra 647 SE to one mole of antibody. The following steps are used to determine the DOS of iFluor™ Ultra 647 SE labeled proteins.
Measure absorption
To measure the absorption spectrum of a dye-protein conjugate, it is recommended to keep the sample concentration in the range of 1-10 µM depending on the extinction coefficient of the dye.
Read OD (absorbance) at 280 nm and dye maximum absorption (ƛmax = 656 nm for iFluor™ Ultra 647 dyes)
For most spectrophotometers, the sample (from the column fractions) need be diluted with de-ionized water so that the OD values are in the range of 0.1 to 0.9. The O.D. (absorbance) at 280 nm is the maximum absorption of protein while 656 nm is the maximum absorption of iFluor™ Ultra 647 SE. To obtain accurate DOS, make sure that the conjugate is free of the non-conjugated dye.
Calculate DOS
You can calculate DOS using our tool by following this link: https://www.aatbio.com/tools/degree-of-labeling-calculator
Spectrum
Product family
| Name | Excitation (nm) | Emission (nm) | Extinction coefficient (cm -1 M -1) | Quantum yield | Correction Factor (260 nm) | Correction Factor (280 nm) |
| iFluor® Ultra 488 succinimidyl ester | 491 | 516 | 750001 | 0.91 | 0.21 | 0.11 |
| iFluor® Ultra 594 succinimidyl ester | 586 | 600 | 1800001 | 0.521 | 0.07 | 0.05 |
| iFluor® Ultra 750 succinimidyl ester | 749 | 773 | 2500001 | 0.321 | 0.04 | 0.05 |
Citations
Authors: Mohale, Mamello and Gundampati, Ravi Kumar and Kumar, Thallapuranam Krishnaswamy Suresh and Heyes, Colin D
Journal: Analytical biochemistry (2022): 114524
Authors: Ji, Tengfei and Ma, Keqiang and Wu, Hongsheng and Cao, Tiansheng
Journal: (2021)
Authors: Masibag, Angelique N and Bergin, Christopher J and Haebe, Joshua R and Zouggar, A{\"\i}cha and Shah, Muhammad S and Sandouka, Tamara and da Silva, Amanda Mendes and Desrochers, Fran{\c{c}}ois M and Fournier-Morin, Aube and Benoit, Yannick D
Journal: Iscience (2021): 103442
Authors: Gerlach, Brennan D and Ampomah, Patrick B and Yurdagul Jr, Arif and Liu, Chuang and Lauring, Max C and Wang, Xiaobo and Kasikara, Canan and Kong, Na and Shi, Jinjun and Tao, Wei and others,
Journal: Cell metabolism (2021): 2445--2463
Authors: Li, Dong and Zhuang, Jie and He, Haisheng and Jiang, Sifan and Banerjee, Amrita and Lu, Yi and Wu, Wei and Mitragotri, Samir and Gan, Li and Qi, Jianping
Journal: ACS applied materials \& interfaces (2017): 42492--42502
References
Authors: Koch, Peter D and Quintana, Jeremy and Ahmed, Maaz and Kohler, Rainer H and Weissleder, Ralph
Journal: Advanced therapeutics (2021)
Authors: Garaeva, Luiza and Kamyshinsky, Roman and Kil, Yury and Varfolomeeva, Elena and Verlov, Nikolai and Komarova, Elena and Garmay, Yuri and Landa, Sergey and Burdakov, Vladimir and Myasnikov, Alexander and Vinnikov, Ilya A and Margulis, Boris and Guzhova, Irina and Kagansky, Alexander and Konevega, Andrey L and Shtam, Tatiana
Journal: Scientific reports (2021): 6489
Authors: Hossain, Farzana and Dohra, Hideo and Yamazaki, Masahito
Journal: Journal of bacteriology (2021)
Authors: Gebhardt, Christian and Lehmann, Martin and Reif, Maria M and Zacharias, Martin and Gemmecker, Gerd and Cordes, Thorben
Journal: Chemphyschem : a European journal of chemical physics and physical chemistry (2021)
Authors: Li, Xiangyu and Fu, Huaxia and Wang, Jing and Liu, Wei and Deng, Hao and Zhao, Peng and Liao, Wei and Yang, Yuchuan and Wei, Hongyuan and Yang, Xia and Chen, Yue
Journal: European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences (2021): 105775