Helixyte™ Green Fluorimetric ssDNA Quantitation Kit
Optimized for Microplate Readers
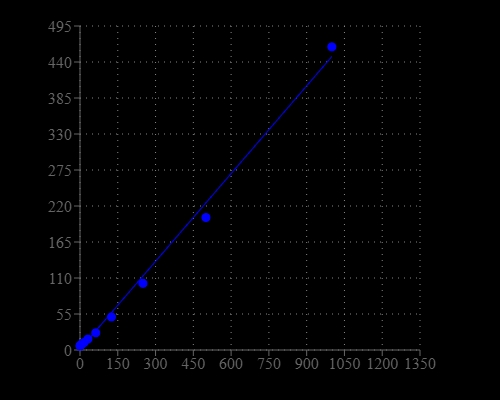
Helixyte™ Green Fluorimetric ssDNA Quantitation Kit is designed to measure single-stranded DNA in an easy and accurate format. The kit has all the essential reagents, including Helixyte™ Green ssDNA reagent, dilution buffer, and prediluted DNA standards. Helixyte™ Green ssDNA reagent is a sensitive fluorescent nucleic acid probe for quantifying oligonucleotides and single-stranded DNA (ssDNA) in solution. It enables researchers to quantify as little as 100 pg/mL oligonucleotide or ssDNA. This sensitivity exceeds that achieved with absorbance methods by more than 10,000-fold. With this kit, as little as 1 ng/mL oligonucleotide or ssDNA might be detected. A few ssDNAs were quantified with this kit, including M13 and ϕX174 viral DNA and denatured calf thymus DNA that had similar sensitivity. Their detection limits were not significantly interfered by the common contaminants in nucleic acid preparations, including salts, urea, ethanol, chloroform, detergents, proteins, ATP, nucleotides and short oligonucleotides of six bases. However, double-stranded DNA (dsDNA) and RNA do interfere with the assay as Helixyte™ Green ssDNA reagent binds to dsDNA and RNA to generate additional fluorescence signals. Helixyte™ Green Fluorimetric ssDNA Quantitation Kit is optimized for quantifying ssDNA with a fluorescence microplate reader.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17623 | 200 Tests | Price |
Spectral properties
| Excitation (nm) | 498 |
| Emission (nm) | 519 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12171501 |
Instrument settings
| Fluorescence microplate reader | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
| Recommended plate | Solid black |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 9, 2026

