Helixyte™ Green Fluorimetric dsDNA Quantitation Kit
High Sensitivity
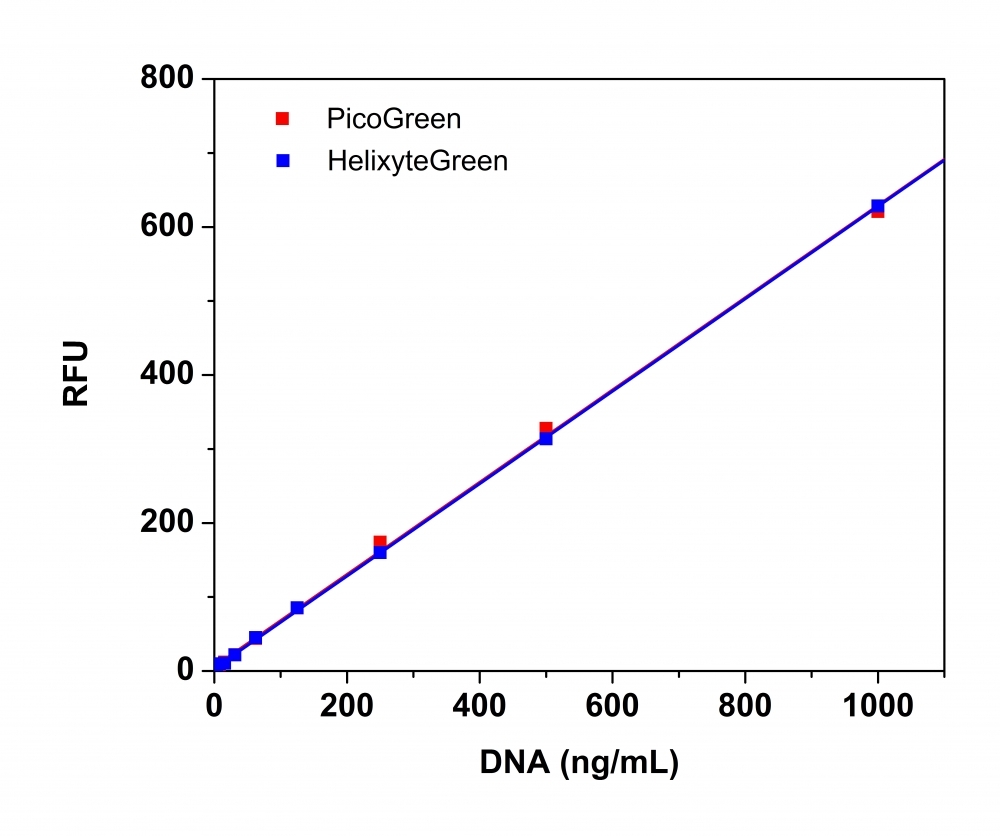
Helixyte™ Green dsDNA Quantitation Assay Kit can be used for selectively detecting as little as 25 pg/ml of dsDNA in the presence of ssDNA, RNA, and free nucleotides. Helixyte™ Green exhibits large fluorescence enhancement upon binding to dsDNA. The assay is linear over three orders of magnitude and has little sequence dependence, allowing you to accurately measure DNA from many sources, including genomic DNA, viral DNA, miniprep DNA, or PCR amplification products. Helixyte™ Green dsDNA Quantitation Assay Kit is a few magnitudes more sensitive than UV absorbance readings. It is specific for dsDNA in the presence of equimolar amounts of RNA. The kit is robust with a mix and read format. It can be used with a bench top fluorometer or a hand-held fluorescence meter (e.g., Qubit fluorometer). This kit is an excellent replacement for Quant-iT™ PicoGreen® dsDNA Assay Kit (Quant-iT™ and PicoGreen® are the trademarks of Invitrogen).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 17651 | 1000 Tests | Price |
Spectral properties
| Excitation (nm) | 502 |
| Emission (nm) | 522 |
Storage, safety and handling
| H-phrase | H303, H313, H340 |
| Hazard symbol | T |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R68 |
| UNSPSC | 41116134 |
Instrument settings
| Spectrofluorometer | |
| Excitation | 490 nm |
| Emission | 525 nm |
| Cutoff | 515 nm |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 20, 2026

