Cell Navigator® Cell Plasma Membrane Staining Kit
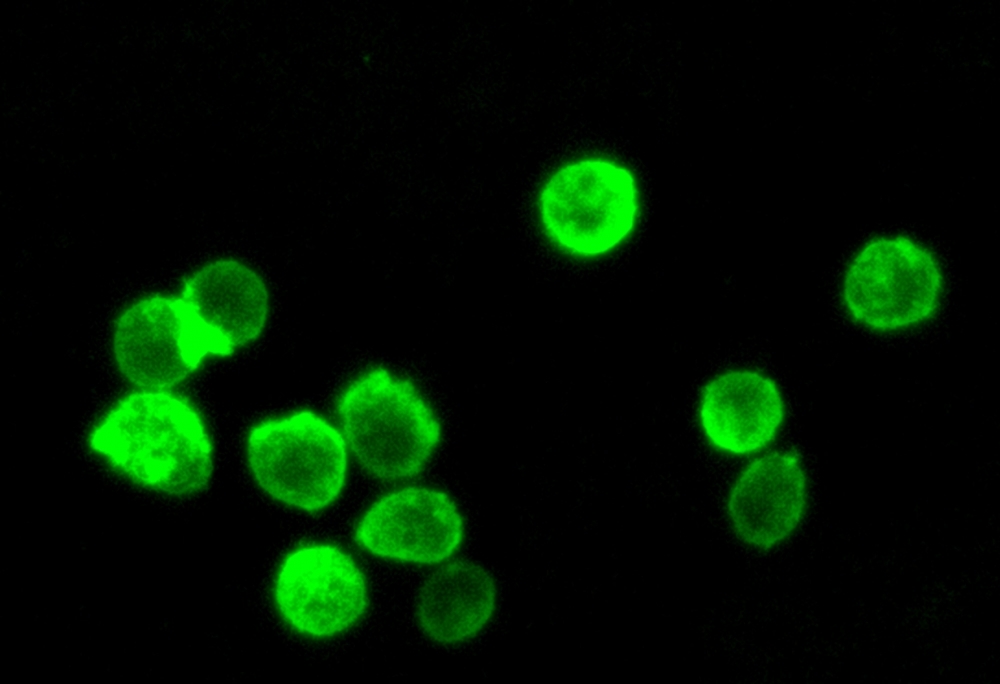
Green Fluorescence
The Cell Navigator® Plasma Membrane Stain Kit provides a fast and uniform labeling of the plasma membrane without the cell-type differences exhibited by lectins. It may be used as a segmentation tool for HCS (high-content screening), as well as to stain cellular plasma membranes for standard fluorescence microscopy. The green fluorescent stain used in the kit survives fixation, but not permeabilization, so it is not suitable for experiments that also involve probing internal targets via antibodies.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 22682 | 500 Tests | Price |
Spectral properties
| Excitation (nm) | 497 |
| Emission (nm) | 505 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| UNSPSC | 12352200 |
Instrument settings
| Fluorescence microscope | |
| Excitation | FITC filter set |
| Emission | FITC filter set |
| Recommended plate | Black wall/clear bottom/Glass slides |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on March 4, 2026

